

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
null
SMILES: CN=CP(C)(O)O
InChI Key: InChIKey=LKLMJGYYKRHTKU-UHFFFAOYSA-N
PDB links: 7 PDB IDs contain this monomer as substructures. 7 PDB IDs contain inhibitors having a similarity of 90% to this monomer.
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Urease subunit alpha [F36L]/beta/gamma (Proteus vulgaris) | BDBM24967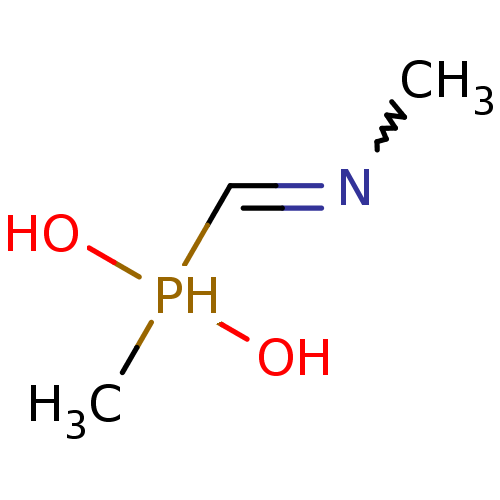 (methyl[(methylamino)methyl]phosphinic acid | organ...) | UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 2.70E+4 | -6.33 | 1.53E+5 | n/a | n/a | n/a | n/a | 7.0 | 30 |
Wroclaw University of Technology | Assay Description Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati... | J Med Chem 51: 5736-44 (2008) Article DOI: 10.1021/jm800570q BindingDB Entry DOI: 10.7270/Q2PR7T91 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Urease subunit alpha/beta/gamma (Bacillus pasteurii) | BDBM24967 (methyl[(methylamino)methyl]phosphinic acid | organ...) | PDB MMDB UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 1.80E+4 | -6.58 | 6.00E+4 | n/a | n/a | n/a | n/a | 7.0 | 30 |
Wroclaw University of Technology | Assay Description Progress curves were obtained by initiation of urease reaction with addition of purified enzyme into assay mixtures containing increasing concentrati... | J Med Chem 51: 5736-44 (2008) Article DOI: 10.1021/jm800570q BindingDB Entry DOI: 10.7270/Q2PR7T91 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||