

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
null
SMILES: C(CN1CCc2ccccc2C1)[C@H]1CC[C@@H](CC1)Nc1ncccn1
InChI Key: InChIKey=BEKYXSCBRPYPCE-IRJFHVNHSA-N
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D(3) dopamine receptor (Homo sapiens (Human)) | BDBM50063278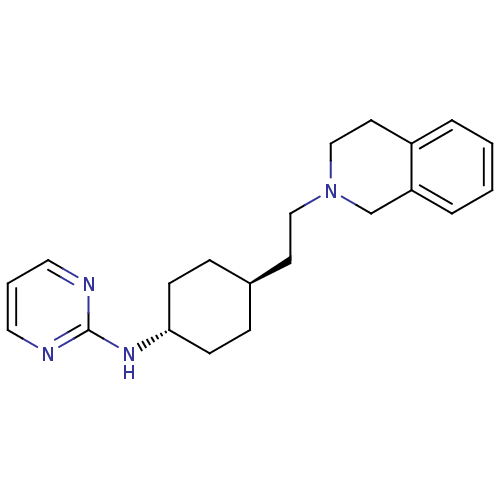 (CHEMBL165440 | {4-[2-(3,4-Dihydro-1H-isoquinolin-2...) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents | Article PubMed | 40 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Warner-Lambert Company Curated by ChEMBL | Assay Description Binding affinity determined by measuring displacement of [3H]-spiperone from cloned Human Dopamine receptor D3 in CHO-K1 cells | J Med Chem 41: 760-71 (1998) Article DOI: 10.1021/jm9707378 BindingDB Entry DOI: 10.7270/Q20G3J97 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| 5-hydroxytryptamine receptor 1A (Rattus norvegicus (rat)) | BDBM50063278 (CHEMBL165440 | {4-[2-(3,4-Dihydro-1H-isoquinolin-2...) | PDB KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents | Article PubMed | 44 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Warner-Lambert Company Curated by ChEMBL | Assay Description The binding affinity of the compound was determined by measuring its ability to displace [3H]-8-OH-DPAT radioligand in 5-hydroxytryptamine 1A recepto... | J Med Chem 41: 760-71 (1998) Article DOI: 10.1021/jm9707378 BindingDB Entry DOI: 10.7270/Q20G3J97 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| D(2) dopamine receptor (Homo sapiens (Human)) | BDBM50063278 (CHEMBL165440 | {4-[2-(3,4-Dihydro-1H-isoquinolin-2...) | PDB Reactome pathway KEGG UniProtKB/SwissProt DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents | Article PubMed | 110 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Warner-Lambert Company Curated by ChEMBL | Assay Description Binding affinity determined by measuring its ability to displace [3H]-N-0437 radioligand in CHO-K1 cells on Cloned Human Dopamine receptor D2 | J Med Chem 41: 760-71 (1998) Article DOI: 10.1021/jm9707378 BindingDB Entry DOI: 10.7270/Q20G3J97 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||