

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
BDBM50458702 CHEMBL4215267
SMILES: COc1ccc2nc(C)c(O[C@@H]3C[C@H](N(C3)C(=O)[C@@H](NC(=O)OC3CCCC3)C(C)(C)C)C(=O)N[C@@]3(C[C@H]3C=C)C(=O)NS(=O)(=O)C3(C)CC3)nc2c1
InChI Key: InChIKey=XMUMASNXHDVGQJ-LCWNRMNXSA-N
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hepatitis C virus serine protease, NS3/NS4A (Hepatitis C virus) | BDBM50458702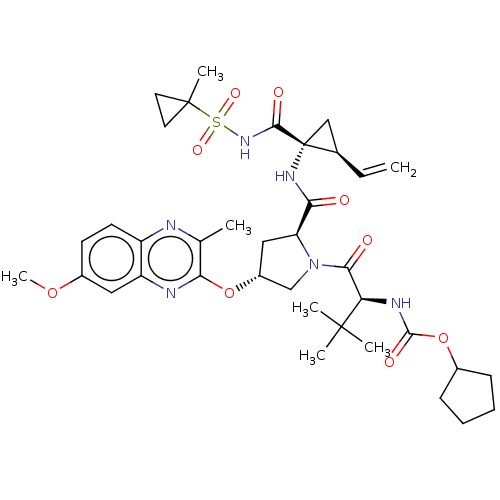 (CHEMBL4215267) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | PC cid PC sid PDB UniChem | PDB Article PubMed | 7.10 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Massachusetts Medical School Curated by ChEMBL | Assay Description Inhibition of wild type HCV genotype 1a NS3/4A protease expressed in Escherichia coli BL21(DE3) using Ac-DE-Dap(QXL 520)-EE-Abu-psi-[COO]AS-C(5-FAMsp... | ACS Med Chem Lett 9: 691-696 (2018) Article DOI: 10.1021/acsmedchemlett.8b00150 BindingDB Entry DOI: 10.7270/Q29C7125 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Hepatitis C virus serine protease, NS3/NS4A (Hepatitis C virus) | BDBM50458702 (CHEMBL4215267) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | PC cid PC sid PDB UniChem | PDB Article PubMed | 10 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Massachusetts Medical School Curated by ChEMBL | Assay Description Inhibition of HCV genotype 1a NS3/4A protease R155K mutant expressed in Escherichia coli BL21(DE3) using Ac-DE-Dap(QXL 520)-EE-Abu-psi-[COO]AS-C(5-FA... | ACS Med Chem Lett 9: 691-696 (2018) Article DOI: 10.1021/acsmedchemlett.8b00150 BindingDB Entry DOI: 10.7270/Q29C7125 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Hepatitis C virus serine protease, NS3/NS4A (Hepatitis C virus) | BDBM50458702 (CHEMBL4215267) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | PC cid PC sid PDB UniChem | PDB Article PubMed | 140 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Massachusetts Medical School Curated by ChEMBL | Assay Description Inhibition of HCV genotype 1a NS3/4A protease D168A mutant expressed in Escherichia coli BL21(DE3) using Ac-DE-Dap(QXL 520)-EE-Abu-psi-[COO]AS-C(5-FA... | ACS Med Chem Lett 9: 691-696 (2018) Article DOI: 10.1021/acsmedchemlett.8b00150 BindingDB Entry DOI: 10.7270/Q29C7125 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||