

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
null
SMILES: [#6]\[#6](-[#6])=[#6]\[#6]-c1cc(-[#6]-2-[#6]-[#6]-c3ccc(-[#8])cc3-[#8]-2)c(-[#6]\[#6]=[#6](/[#6])-[#6])c(-[#8])c1-[#8]
InChI Key: InChIKey=QXHVECWDOBLWPW-UHFFFAOYSA-N
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alpha-glucosidase MAL62 (Saccharomyces cerevisiae) | BDBM50486892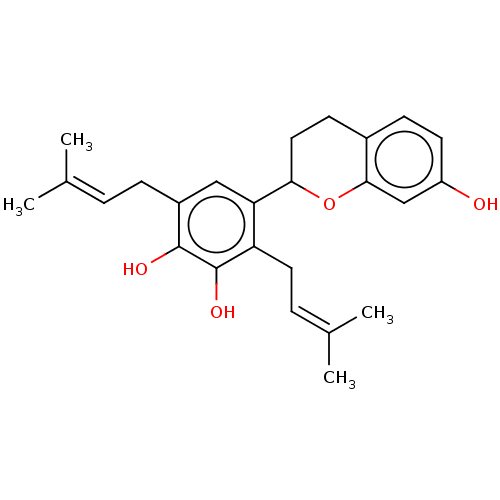 (CHEMBL2235108) | Reactome pathway UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article | n/a | n/a | 1.20E+10 | n/a | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Inhibition of baker's yeast alpha-glucosidase using p-nitrophenyl-alpha-D-glucopyranoside as substrate by spectrophotometry | Citation and Details Article DOI: 10.1007/s00044-011-9938-0 BindingDB Entry DOI: 10.7270/Q29W0JBG | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||