

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
BDBM50537586 CHEMBL4641917
SMILES: CS(=O)(=O)Nc1cccc(c1)S(=O)(=O)NC(=O)c1c(-c2ccc[nH]c2=O)c2ccc(Cl)cc2n1Cc1ccnc(N)c1
InChI Key: InChIKey=JMGANSJPVOEXRZ-UHFFFAOYSA-N
Data: 1 IC50
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hepatitis C virus NS5B RNA-dependent RNA polymerase (Hepatitis C virus) | BDBM50537586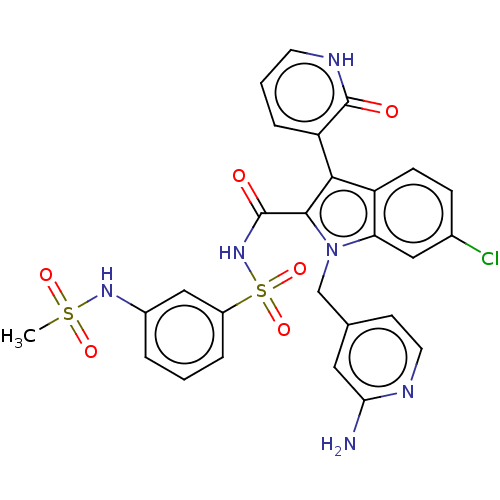 (CHEMBL4641917) | UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 39 | n/a | n/a | n/a | n/a | n/a | n/a |
Institute of Pharmaceutical Education and Research Curated by ChEMBL | Assay Description Inhibitory activity against HIV-1 Mutant Reverse transcriptase P236L | Eur J Med Chem 164: 576-601 (2019) Article DOI: 10.1007/s00044-005-0131-1 BindingDB Entry DOI: 10.7270/Q2RR224D | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||