BDBM92285 CS279
SMILES: ONC(=O)[C@H]1COC(=N1)c1ccc(F)cc1
InChI Key: InChIKey=KUIMFZQMQKZITC-MRVPVSSYSA-N
Activity Spreadsheet -- Enzyme Inhibition Constant Data from BindingDB
 Found 3 hits for monomerid = 92285
Found 3 hits for monomerid = 92285 Target/Host
(Institution) | Ligand | Target/Host
Links | Ligand
Links | Trg + Lig
Links | Ki
nM | ΔG°
kcal/mole | IC50
nM | Kd
nM | EC50/IC50
nM | koff
s-1 | kon
M-1s-1 | pH | Temp
°C |
|---|
UDP-3-O-Acyl-GlcNAc Deacetylase (LpxC)
(Aquifex aeolicus) | BDBM92285
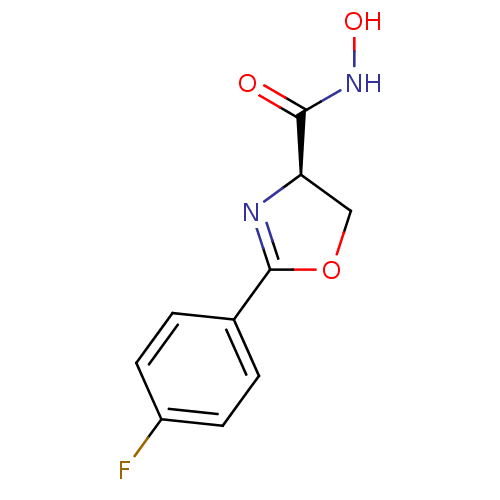
(CS279)Show InChI InChI=1S/C10H9FN2O3/c11-7-3-1-6(2-4-7)10-12-8(5-16-10)9(14)13-15/h1-4,8,15H,5H2,(H,13,14)/t8-/m1/s1 | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Similars
| PubMed
| n/a | n/a | n/a | 1.11E+4 | n/a | n/a | n/a | 7.0 | 25 |
CSAR
| Assay Description
Thermofluor |
CSAR 1: (2012)
|
More data for this
Ligand-Target Pair | |
UDP-3-O-acyl-GlcNAc deacetylase (LpxC)
(Pseudomonas aeruginosa) | BDBM92285

(CS279)Show InChI InChI=1S/C10H9FN2O3/c11-7-3-1-6(2-4-7)10-12-8(5-16-10)9(14)13-15/h1-4,8,15H,5H2,(H,13,14)/t8-/m1/s1 | PDB
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Similars
| D3R
| n/a | n/a | n/a | 1.11E+4 | n/a | n/a | n/a | n/a | n/a |
D3R
| Assay Description
Thermofluor_Method1 |
D3R 224: (2015)
BindingDB Entry DOI: 10.7270/Q2HM57BR |
More data for this
Ligand-Target Pair | |
UDP-3-O-Acyl-GlcNAc Deacetylase (LpxC)
(Aquifex aeolicus) | BDBM92285

(CS279)Show InChI InChI=1S/C10H9FN2O3/c11-7-3-1-6(2-4-7)10-12-8(5-16-10)9(14)13-15/h1-4,8,15H,5H2,(H,13,14)/t8-/m1/s1 | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Similars
| PubMed
| n/a | n/a | n/a | 1.48E+4 | n/a | n/a | n/a | 7.0 | 25 |
CSAR
| Assay Description
ITC |
CSAR 1: (2012)
|
More data for this
Ligand-Target Pair | |


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data