

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell division protein FtsZ (Bacillus subtilis) | BDBM50581021 (CHEMBL5076899) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 9.30E+3 | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysis | Citation and Details Article DOI: 10.1021/acs.jmedchem.0c02207 BindingDB Entry DOI: 10.7270/Q2TT4VTZ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cell division protein FtsZ (Bacillus subtilis) | BDBM50581039 (CHEMBL5093170) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 1.00E+6 | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Displacement of free fluorescent probe from inter domain cleft of stabilized Bacillus subtilis FtsZ assessed as dissociation constant in presence of ... | Citation and Details Article DOI: 10.1021/acs.jmedchem.0c02207 BindingDB Entry DOI: 10.7270/Q2TT4VTZ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cell division protein FtsZ (Bacillus subtilis) | BDBM50581023 (CHEMBL5079700) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 9.50E+3 | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysis | Citation and Details Article DOI: 10.1021/acs.jmedchem.0c02207 BindingDB Entry DOI: 10.7270/Q2TT4VTZ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cell division protein FtsZ (Bacillus subtilis) | BDBM50581024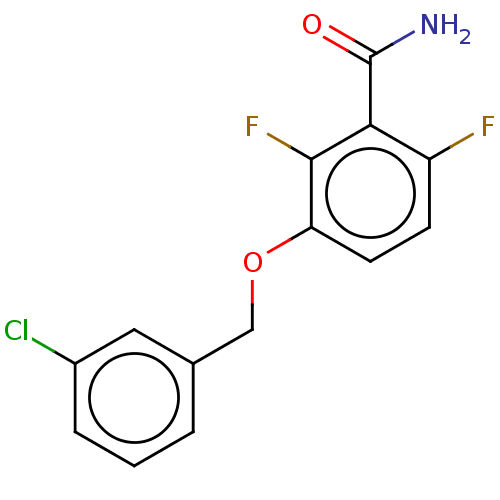 (CHEMBL5092442) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid PDB UniChem | Article PubMed | n/a | n/a | n/a | 1.30E+3 | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysis | Citation and Details Article DOI: 10.1021/acs.jmedchem.0c02207 BindingDB Entry DOI: 10.7270/Q2TT4VTZ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cell division protein FtsZ (Bacillus subtilis) | BDBM50581025 (CHEMBL5078275) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 7.10E+4 | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysis | Citation and Details Article DOI: 10.1021/acs.jmedchem.0c02207 BindingDB Entry DOI: 10.7270/Q2TT4VTZ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cell division protein FtsZ (Bacillus subtilis) | BDBM50581026 (CHEMBL5080731) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid PDB UniChem | Article PubMed | n/a | n/a | n/a | 700 | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysis | Citation and Details Article DOI: 10.1021/acs.jmedchem.0c02207 BindingDB Entry DOI: 10.7270/Q2TT4VTZ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cell division protein FtsZ (Bacillus subtilis) | BDBM50581027 (CHEMBL5081697) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 6.70E+3 | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysis | Citation and Details Article DOI: 10.1021/acs.jmedchem.0c02207 BindingDB Entry DOI: 10.7270/Q2TT4VTZ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cell division protein FtsZ (Bacillus subtilis) | BDBM50581028 (CHEMBL5090219) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 5.90E+3 | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysis | Citation and Details Article DOI: 10.1021/acs.jmedchem.0c02207 BindingDB Entry DOI: 10.7270/Q2TT4VTZ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cell division protein FtsZ (Bacillus subtilis) | BDBM50581029 (CHEMBL5085966) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 1.50E+3 | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysis | Citation and Details Article DOI: 10.1021/acs.jmedchem.0c02207 BindingDB Entry DOI: 10.7270/Q2TT4VTZ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cell division protein FtsZ (Bacillus subtilis) | BDBM50581030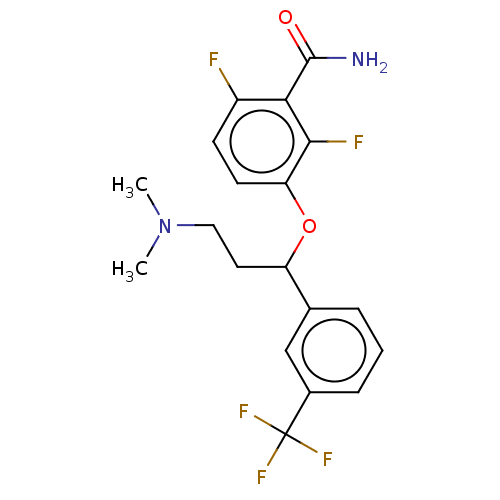 (CHEMBL5091172) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 1.45E+5 | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysis | Citation and Details Article DOI: 10.1021/acs.jmedchem.0c02207 BindingDB Entry DOI: 10.7270/Q2TT4VTZ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cell division protein FtsZ (Bacillus subtilis) | BDBM50581031 (CHEMBL5074266) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 1.90E+3 | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysis | Citation and Details Article DOI: 10.1021/acs.jmedchem.0c02207 BindingDB Entry DOI: 10.7270/Q2TT4VTZ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cell division protein FtsZ (Bacillus subtilis) | BDBM50581032 (CHEMBL5076150) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 1.90E+3 | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysis | Citation and Details Article DOI: 10.1021/acs.jmedchem.0c02207 BindingDB Entry DOI: 10.7270/Q2TT4VTZ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cell division protein FtsZ (Bacillus subtilis) | BDBM50581033 (CHEMBL5076408) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 1.10E+3 | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysis | Citation and Details Article DOI: 10.1021/acs.jmedchem.0c02207 BindingDB Entry DOI: 10.7270/Q2TT4VTZ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cell division protein FtsZ (Bacillus subtilis) | BDBM50581034 (CHEMBL5085404) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 200 | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysis | Citation and Details Article DOI: 10.1021/acs.jmedchem.0c02207 BindingDB Entry DOI: 10.7270/Q2TT4VTZ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cell division protein FtsZ (Bacillus subtilis) | BDBM50581035 (CHEMBL5089927) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 3.80E+3 | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysis | Citation and Details Article DOI: 10.1021/acs.jmedchem.0c02207 BindingDB Entry DOI: 10.7270/Q2TT4VTZ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cell division protein FtsZ (Bacillus subtilis) | BDBM50581036 (CHEMBL5092196) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 7.10E+3 | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysis | Citation and Details Article DOI: 10.1021/acs.jmedchem.0c02207 BindingDB Entry DOI: 10.7270/Q2TT4VTZ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cell division protein FtsZ (Bacillus subtilis) | BDBM50581037 (CHEMBL5081559) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 1.50E+4 | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysis | Citation and Details Article DOI: 10.1021/acs.jmedchem.0c02207 BindingDB Entry DOI: 10.7270/Q2TT4VTZ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cell division protein FtsZ (Bacillus subtilis) | BDBM50079356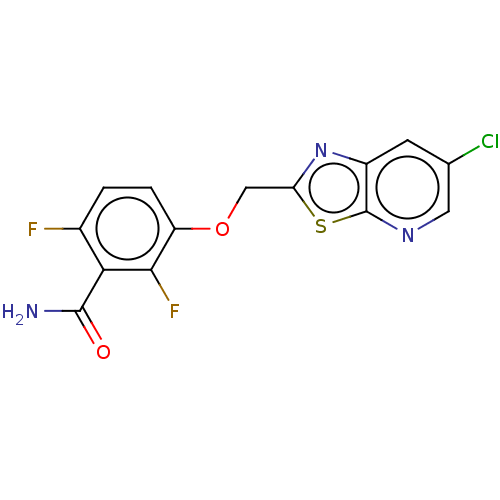 (CHEMBL511201) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | n/a | <100 | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Displacement of free fluorescent probe from inter domain cleft of stabilized Bacillus subtilis FtsZ assessed as dissociation constant in presence of ... | Citation and Details Article DOI: 10.1021/acs.jmedchem.0c02207 BindingDB Entry DOI: 10.7270/Q2TT4VTZ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cell division protein FtsZ (Bacillus subtilis) | BDBM50581038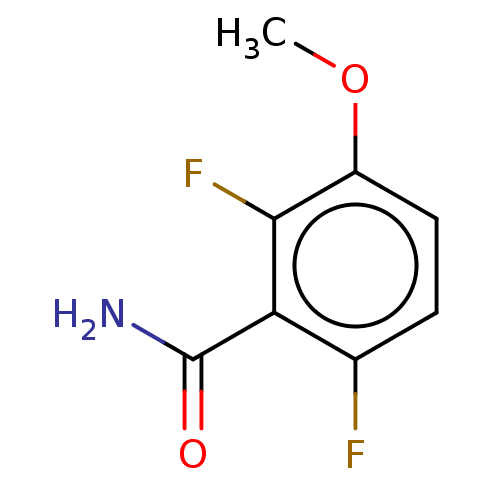 (CHEMBL453452) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid PDB UniChem | Article PubMed | n/a | n/a | n/a | 6.70E+5 | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Displacement of free fluorescent probe from inter domain cleft of stabilized Bacillus subtilis FtsZ assessed as dissociation constant in presence of ... | Citation and Details Article DOI: 10.1021/acs.jmedchem.0c02207 BindingDB Entry DOI: 10.7270/Q2TT4VTZ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cell division protein FtsZ (Bacillus subtilis) | BDBM50581022 (CHEMBL5090793) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 8.00E+3 | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Binding affinity to Bacillus subtilis FtsZ polymer assessed as dissociation constant by fluorescence anisotropy analysis | Citation and Details Article DOI: 10.1021/acs.jmedchem.0c02207 BindingDB Entry DOI: 10.7270/Q2TT4VTZ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||