C-C chemokine receptor type 5
(Homo sapiens (Human)) | BDBM50257641
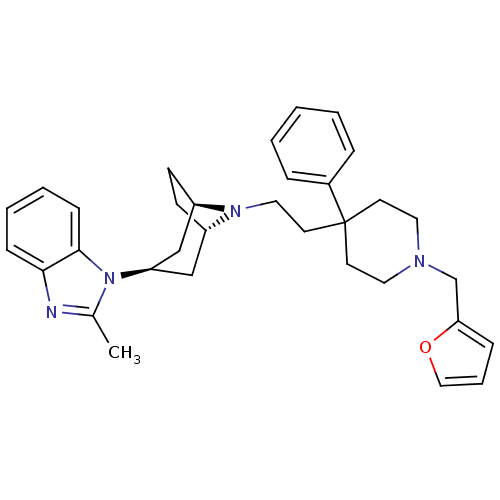
(1-(8-(2-(1-(furan-2-ylmethyl)-4-phenylpiperidin-4-...)Show SMILES Cc1nc2ccccc2n1[C@@H]1C[C@@H]2CC[C@H](C1)N2CCC1(CCN(Cc2ccco2)CC1)c1ccccc1 |r,TLB:9:10:17:13.14| Show InChI InChI=1S/C33H40N4O/c1-25-34-31-11-5-6-12-32(31)37(25)29-22-27-13-14-28(23-29)36(27)20-17-33(26-8-3-2-4-9-26)15-18-35(19-16-33)24-30-10-7-21-38-30/h2-12,21,27-29H,13-20,22-24H2,1H3/t27-,28+,29+ | PDB
KEGG
UniProtKB/SwissProt
B.MOAD
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| CHEMBL
PC cid
PC sid
UniChem
Patents
Similars
| Article
PubMed
| n/a | n/a | 3 | n/a | n/a | n/a | n/a | n/a | n/a |