Peroxisome proliferator-activated receptor alpha
(Homo sapiens (Human)) | BDBM50275536
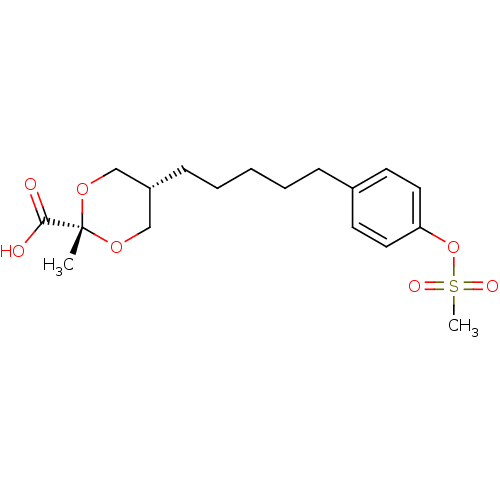
(CHEMBL485942 | cis-2-methyl-5-(5-(4-(methylsulfony...)Show SMILES C[C@]1(OC[C@H](CCCCCc2ccc(OS(C)(=O)=O)cc2)CO1)C(O)=O |r,wU:4.4,1.24,wD:1.0,(9.11,1.64,;7.58,1.63,;7.58,3.17,;6.25,3.94,;4.93,3.18,;3.59,3.95,;2.26,3.17,;.92,3.94,;-.41,3.17,;-1.74,3.94,;-3.07,3.17,;-3.07,1.63,;-4.41,.86,;-5.75,1.63,;-7.08,.86,;-7.08,-.67,;-7.08,-2.21,;-5.54,-.67,;-8.62,-.67,;-5.74,3.17,;-4.42,3.94,;4.92,1.63,;6.24,.87,;7.57,.1,;8.89,-.68,;6.23,-.66,)| Show InChI InChI=1S/C18H26O7S/c1-18(17(19)20)23-12-15(13-24-18)7-5-3-4-6-14-8-10-16(11-9-14)25-26(2,21)22/h8-11,15H,3-7,12-13H2,1-2H3,(H,19,20)/t15-,18+ | PDB
UniProtKB/SwissProt
antibodypedia
GoogleScholar
AffyNet 
| CHEMBL
PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | n/a | n/a | 2.50E+3 | n/a | n/a | n/a | n/a |