C-X-C chemokine receptor type 3
(Homo sapiens (Human)) | BDBM50200678
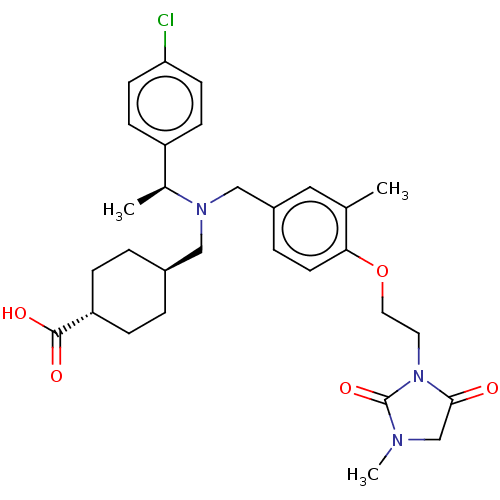
(CHEMBL3970774)Show SMILES C[C@H](N(C[C@H]1CC[C@@H](CC1)C(O)=O)Cc1ccc(OCCN2C(=O)CN(C)C2=O)c(C)c1)c1ccc(Cl)cc1 |r,wU:1.0,4.3,wD:7.10,(33.96,-33.61,;33.96,-32.07,;35.32,-31.35,;35.32,-29.8,;36.72,-29.07,;36.72,-27.54,;38.13,-26.81,;39.44,-27.62,;39.44,-29.16,;38.03,-29.89,;40.79,-26.9,;42.1,-27.71,;40.79,-25.36,;36.59,-32.15,;36.59,-33.69,;35.23,-34.42,;35.23,-35.96,;36.5,-36.76,;36.5,-38.3,;37.76,-39.11,;37.76,-40.65,;36.4,-41.52,;36.4,-43.06,;37.61,-44.01,;34.92,-43.5,;34.05,-42.22,;32.51,-42.22,;35,-41,;34.56,-39.52,;37.86,-36.03,;39.17,-36.83,;37.86,-34.49,;32.65,-31.27,;31.29,-31.99,;30.04,-31.19,;30.04,-29.65,;28.73,-28.83,;31.38,-28.92,;32.65,-29.72,)| Show InChI InChI=1S/C30H38ClN3O5/c1-20-16-23(6-13-27(20)39-15-14-34-28(35)19-32(3)30(34)38)18-33(21(2)24-9-11-26(31)12-10-24)17-22-4-7-25(8-5-22)29(36)37/h6,9-13,16,21-22,25H,4-5,7-8,14-15,17-19H2,1-3H3,(H,36,37)/t21-,22-,25-/m0/s1 | PDB
UniProtKB/SwissProt
antibodypedia
GoogleScholar
AffyNet 
| CHEMBL
PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | 99 | n/a | n/a | n/a | n/a | n/a | n/a |