Diacylglycerol O-acyltransferase 2
(Homo sapiens (Human)) | BDBM530628
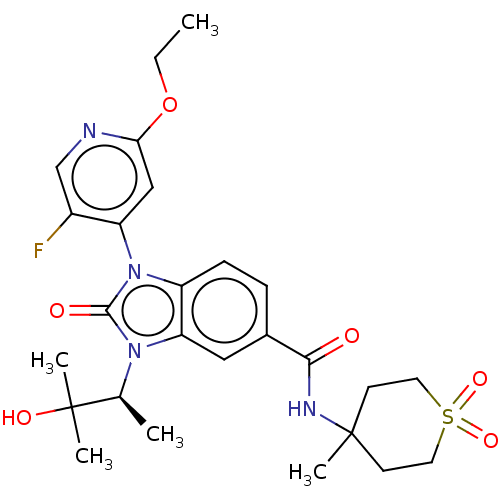
(WO2022076495, EXAMPLE 69a)Show SMILES CCOc1cc(c(F)cn1)-n1c2ccc(cc2n([C@@H](C)C(C)(C)O)c1=O)C(=O)NC1(C)CCS(=O)(=O)CC1 |r,wU:18.20,(-4.38,-5.29,;-2.87,-4.97,;-1.84,-6.12,;-.33,-5.8,;.14,-4.33,;1.65,-4.01,;2.68,-5.16,;4.19,-4.84,;2.2,-6.62,;.7,-6.94,;2.13,-2.55,;3.59,-2.07,;4.92,-2.84,;6.26,-2.07,;6.26,-.53,;4.92,.24,;3.59,-.53,;2.13,-.06,;1.65,1.41,;.14,1.73,;2.68,2.55,;4.19,2.23,;3.16,1.09,;2.2,4.02,;1.22,-1.3,;-.32,-1.3,;7.59,.24,;7.59,1.78,;8.93,-.53,;10.26,.24,;10.79,-1.21,;9.27,1.42,;9.8,2.86,;11.31,3.13,;10.79,4.58,;12.65,3.9,;12.3,1.95,;11.78,.5,)| | Reactome pathway
KEGG
UniProtKB/SwissProt
antibodypedia
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
| WIPO WO2022076495
| n/a | n/a | 9.70 | n/a | n/a | n/a | n/a | n/a | n/a |