Prostaglandin E synthase
(Homo sapiens (Human)) | BDBM118846
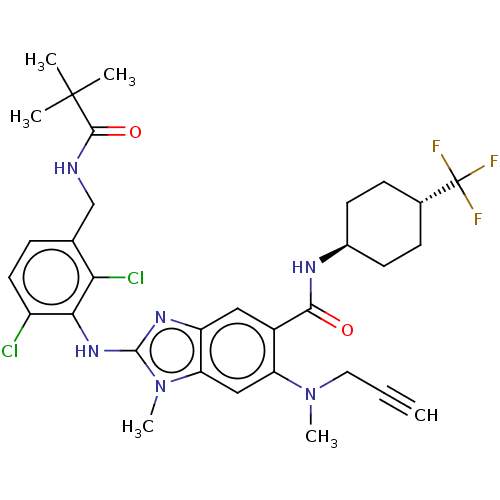
(US8674113, 103)Show SMILES CN(CC#C)c1cc2n(C)c(Nc3c(Cl)ccc(CNC(=O)C(C)(C)C)c3Cl)nc2cc1C(=O)N[C@H]1CC[C@@H](CC1)C(F)(F)F |r,wU:38.44,wD:35.37,(-5.96,.77,;-4.62,,;-4.62,-1.54,;-5.96,-2.31,;-7.29,-3.08,;-3.29,.77,;-1.95,,;-.62,.77,;.84,.29,;.84,-1.25,;1.75,1.54,;3.29,1.54,;4.62,.77,;5.96,1.54,;5.96,3.08,;7.29,.77,;7.29,-.77,;5.96,-1.54,;5.96,-3.08,;7.29,-3.85,;8.62,-3.08,;8.62,-1.54,;9.96,-3.85,;11.29,-3.08,;9.96,-5.39,;11.29,-4.62,;4.62,-.77,;3.29,-1.54,;.84,2.79,;-.62,2.31,;-1.95,3.08,;-3.29,2.31,;-4.62,3.08,;-4.62,4.62,;-5.96,2.31,;-7.29,3.08,;-7.29,4.62,;-8.62,5.39,;-9.96,4.62,;-9.96,3.08,;-8.62,2.31,;-11.29,5.39,;-12.78,4.99,;-12.62,6.16,;-11.29,6.93,)| Show InChI InChI=1S/C32H37Cl2F3N6O2/c1-7-14-42(5)24-16-25-23(15-21(24)28(44)39-20-11-9-19(10-12-20)32(35,36)37)40-30(43(25)6)41-27-22(33)13-8-18(26(27)34)17-38-29(45)31(2,3)4/h1,8,13,15-16,19-20H,9-12,14,17H2,2-6H3,(H,38,45)(H,39,44)(H,40,41)/t19-,20- | PDB
MMDB
Reactome pathway
KEGG
UniProtKB/SwissProt
B.MOAD
antibodypedia
GoogleScholar
AffyNet 
| PC cid
PC sid
UniChem
Similars
| US Patent
| n/a | n/a | 1 | n/a | n/a | n/a | n/a | 7.4 | 25 |