Perforin-1
(Homo sapiens (Human)) | BDBM50361435
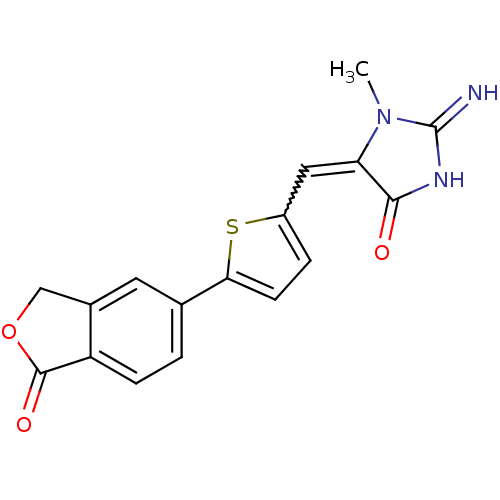
(CHEMBL1934399)Show SMILES CN1C(=N)NC(=O)\C1=C/c1ccc(s1)-c1ccc2C(=O)OCc2c1 |w:9.9| Show InChI InChI=1S/C17H13N3O3S/c1-20-13(15(21)19-17(20)18)7-11-3-5-14(24-11)9-2-4-12-10(6-9)8-23-16(12)22/h2-7H,8H2,1H3,(H2,18,19,21)/b13-7+ | NCI pathway
KEGG
UniProtKB/SwissProt
antibodypedia
GoogleScholar
AffyNet 
| CHEMBL
PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | 2.75E+3 | n/a | n/a | n/a | n/a | n/a | n/a |