 Found 2 hits in this display
Found 2 hits in this display Target/Host
(Institution) | Ligand | Target/Host
Links | Ligand
Links | Trg + Lig
Links | Ki
nM | ΔG°
kJ/mole | IC50
nM | Kd
nM | EC50/IC50
nM | koff
s-1 | kon
M-1s-1 | pH | Temp
°C |
|---|
Polymerase acidic protein
(Hepatitis C virus) | BDBM50125805
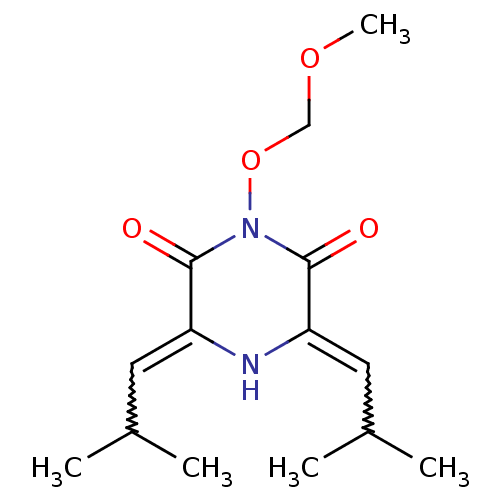
((3Z)-5-isobutyl-1-(methoxymethoxy)-3-(2-methylprop...)Show SMILES COCOn1c(=O)c(=CC(C)C)[nH]c(=CC(C)C)c1=O |w:8.8,14.14| Show InChI InChI=1S/C14H22N2O4/c1-9(2)6-11-13(17)16(20-8-19-5)14(18)12(15-11)7-10(3)4/h6-7,9-10,15H,8H2,1-5H3 | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| CHEMBL
PC cid
PC sid
UniChem
Similars
| PubMed
| n/a | n/a | >2.00E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories
Curated by ChEMBL
| Assay Description
Inhibition of cap 1 ALMV primed Influenza transcriptase |
J Org Chem 66: 5504-16 (2001)
BindingDB Entry DOI: 10.7270/Q298882D |
More data for this
Ligand-Target Pair | |
Polymerase acidic protein
(Hepatitis C virus) | BDBM50125805

((3Z)-5-isobutyl-1-(methoxymethoxy)-3-(2-methylprop...)Show SMILES COCOn1c(=O)c(=CC(C)C)[nH]c(=CC(C)C)c1=O |w:8.8,14.14| Show InChI InChI=1S/C14H22N2O4/c1-9(2)6-11-13(17)16(20-8-19-5)14(18)12(15-11)7-10(3)4/h6-7,9-10,15H,8H2,1-5H3 | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| CHEMBL
PC cid
PC sid
UniChem
Similars
| Article
PubMed
| n/a | n/a | >3.55E+5 | n/a | n/a | n/a | n/a | n/a | n/a |
Roche Discovery Welwyn
Curated by ChEMBL
| Assay Description
Inhibitory concentration against cap-dependent endonuclease activity of influenza virus RNP |
J Med Chem 46: 1153-64 (2003)
Article DOI: 10.1021/jm020334u
BindingDB Entry DOI: 10.7270/Q22J6B6V |
More data for this
Ligand-Target Pair | |