Polymerase acidic protein
(Hepatitis C virus) | BDBM50385498
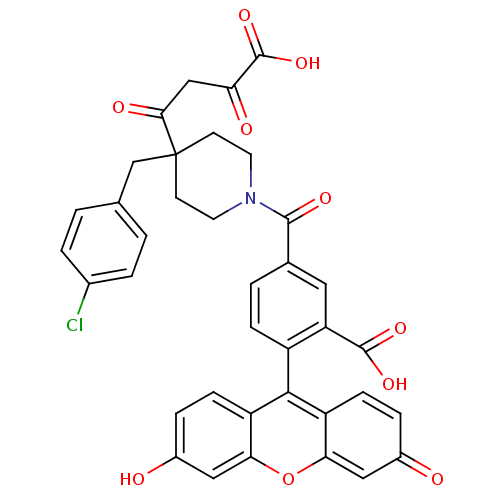
(CHEMBL2040551)Show SMILES OC(=O)C(=O)CC(=O)C1(Cc2ccc(Cl)cc2)CCN(CC1)C(=O)c1ccc(c(c1)C(O)=O)-c1c2ccc(O)cc2oc2cc(=O)ccc12 |(18.46,-2.91,;17.13,-3.68,;17.13,-5.22,;15.79,-2.91,;15.79,-1.37,;14.46,-3.68,;13.13,-2.91,;13.13,-1.37,;11.79,-3.68,;12.73,-4.9,;12.73,-6.44,;11.4,-7.21,;11.4,-8.75,;12.73,-9.52,;12.73,-11.06,;14.06,-8.75,;14.06,-7.21,;10.86,-4.9,;9.33,-4.7,;8.74,-3.28,;9.68,-2.06,;11.2,-2.26,;7.21,-3.08,;6.28,-4.3,;6.62,-1.66,;7.56,-.43,;6.97,.99,;5.44,1.19,;4.51,-.03,;5.1,-1.45,;2.97,-.03,;1.54,.56,;3.17,-1.56,;4.86,2.61,;5.79,3.83,;7.32,3.63,;8.26,4.86,;7.67,6.28,;8.61,7.5,;6.14,6.48,;5.2,5.26,;3.68,5.46,;2.74,4.24,;1.21,4.44,;.28,3.22,;-1.25,3.42,;.86,1.79,;2.39,1.59,;3.33,2.81,)| Show InChI InChI=1S/C37H28ClNO10/c38-22-4-1-20(2-5-22)19-37(32(43)18-29(42)36(47)48)11-13-39(14-12-37)34(44)21-3-8-25(28(15-21)35(45)46)33-26-9-6-23(40)16-30(26)49-31-17-24(41)7-10-27(31)33/h1-10,15-17,40H,11-14,18-19H2,(H,45,46)(H,47,48) | PDB
MMDB
KEGG
UniProtKB/SwissProt
B.MOAD
GoogleScholar
AffyNet 
| CHEMBL
KEGG
PC cid
PC sid
UniChem
| Article
PubMed
| n/a | n/a | n/a | 350 | n/a | n/a | n/a | n/a | n/a |