

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sodium-dependent dopamine transporter (Rattus norvegicus (rat)) | BDBM50129592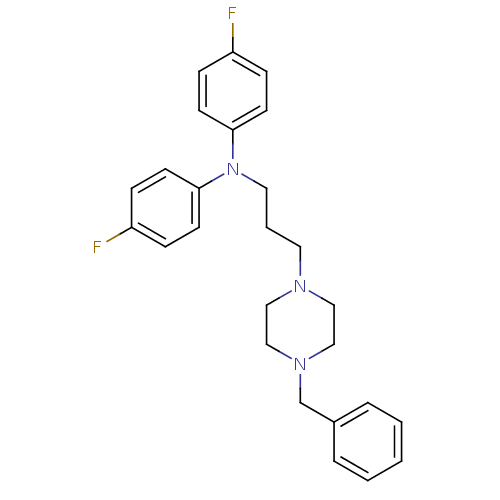 (CHEMBL87841 | N-(3-(4-benzylpiperazin-1-yl)propyl)...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem | Article PubMed | 27.6 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
NIDA-IRP Curated by ChEMBL | Assay Description Binding affinity against dopamine transporter was determined by the displacement of [3H]WIN-35428 radioligand in rat brain | J Med Chem 46: 2589-98 (2003) Article DOI: 10.1021/jm030008u BindingDB Entry DOI: 10.7270/Q2H70GJN | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Sodium-dependent dopamine transporter (Rattus norvegicus (rat)) | BDBM50129592 (CHEMBL87841 | N-(3-(4-benzylpiperazin-1-yl)propyl)...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem | Article PubMed | n/a | n/a | 29.7 | n/a | n/a | n/a | n/a | n/a | n/a |
National Institute on Drug Abuse Curated by ChEMBL | Assay Description Displacement of [3H]WIN 35428 from DAT in Sprague-Dawley rat striatum | J Med Chem 47: 6128-36 (2004) Article DOI: 10.1021/jm049670w BindingDB Entry DOI: 10.7270/Q2JH3KNF | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||