

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UDP-3-O-acyl-N-acetylglucosamine deacetylase (Escherichia coli) | BDBM50074941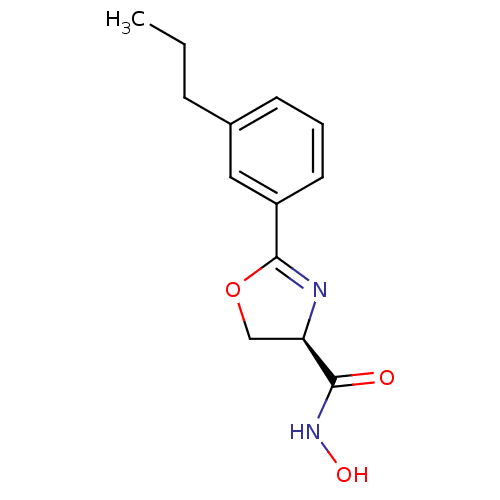 ((R)-2-(3-Propyl-phenyl)-4,5-dihydro-oxazole-4-carb...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | n/a | n/a | 50 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of UDP-3-O-[R-3-hydroxymyristoyl]-GlcNAc deacetylase using direct deacetylase assay (DEACET) in E. coli strain JB 1104 | Bioorg Med Chem Lett 9: 313-8 (1999) BindingDB Entry DOI: 10.7270/Q2CJ8CNN | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| UDP-3-O-acyl-N-acetylglucosamine deacetylase (Escherichia coli) | BDBM50074941 ((R)-2-(3-Propyl-phenyl)-4,5-dihydro-oxazole-4-carb...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | n/a | n/a | 50 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of UDP-3-O-[R-3-hydroxymyristoyl]-GlcNAc deacetylase using coupled radiochemical assay (WAVE) in E. coli strain JB 1104 | Bioorg Med Chem Lett 9: 313-8 (1999) BindingDB Entry DOI: 10.7270/Q2CJ8CNN | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||