

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D(2) dopamine receptor (Rattus norvegicus (rat)) | BDBM50061648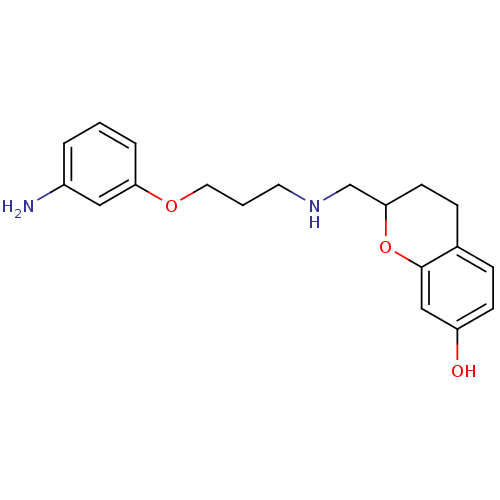 (2-{[3-(3-Amino-phenoxy)-propylamino]-methyl}-chrom...) | PDB Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 0.900 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Wyeth-Ayerst Research Laboratories Curated by ChEMBL | Assay Description In vitro binding affinity to rat striatal Dopamine receptor D2 was determined using the agonist [3H]quinpirole to label the high affinity state (D2 h... | J Med Chem 40: 4235-56 (1998) Article DOI: 10.1021/jm9703653 BindingDB Entry DOI: 10.7270/Q2KP82TD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| 5-hydroxytryptamine receptor 1A (Rattus norvegicus (rat)) | BDBM50061648 (2-{[3-(3-Amino-phenoxy)-propylamino]-methyl}-chrom...) | PDB KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 2.60 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Wyeth-Ayerst Research Laboratories Curated by ChEMBL | Assay Description In vitro binding affinity to rat hippocampal 5-hydroxytryptamine 1A receptor was determined using [3H]8-OH-DPAT as radioligand | J Med Chem 40: 4235-56 (1998) Article DOI: 10.1021/jm9703653 BindingDB Entry DOI: 10.7270/Q2KP82TD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| D(2) dopamine receptor (Rattus norvegicus (rat)) | BDBM50061648 (2-{[3-(3-Amino-phenoxy)-propylamino]-methyl}-chrom...) | PDB Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 16 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Wyeth-Ayerst Research Laboratories Curated by ChEMBL | Assay Description In vitro binding affinity to rat striatal Dopamine receptor D2 was determined using the antagonist [3H]spiperone + GTP to label the low affinity stat... | J Med Chem 40: 4235-56 (1998) Article DOI: 10.1021/jm9703653 BindingDB Entry DOI: 10.7270/Q2KP82TD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||