

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4-aminobutyrate aminotransferase, mitochondrial (Sus scrofa) | BDBM50049260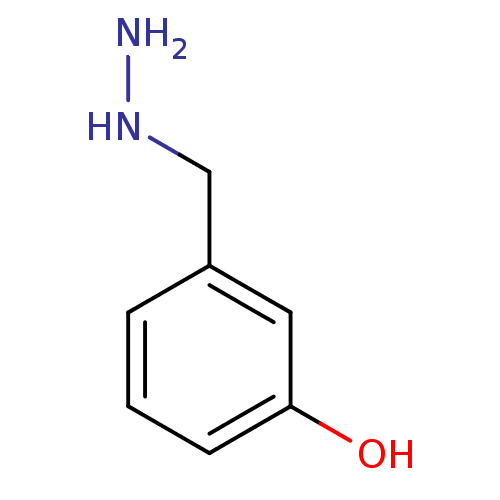 (3-Hydrazinomethyl-phenol | CHEMBL352205) | PDB MMDB Reactome pathway UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid UniChem Patents | Article PubMed | n/a | n/a | n/a | 460 | n/a | n/a | n/a | n/a | n/a |
Northwestern University Curated by ChEMBL | Assay Description The Dissociation Constant of the compound for Gamma-amino-N-butyrate transaminase | J Med Chem 39: 686-94 (1996) Article DOI: 10.1021/jm950437v BindingDB Entry DOI: 10.7270/Q2CJ8CKR | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| 4-aminobutyrate aminotransferase, mitochondrial (Sus scrofa) | BDBM50049259 (CHEMBL160520 | Methyl-hydrazine) | PDB MMDB Reactome pathway UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL MCE MMDB PC cid PC sid UniChem Patents | Article PubMed | n/a | n/a | n/a | 2.80E+3 | n/a | n/a | n/a | n/a | n/a |
Northwestern University Curated by ChEMBL | Assay Description The Dissociation Constant of the compound for Gamma-amino-N-butyrate transaminase | J Med Chem 39: 686-94 (1996) Article DOI: 10.1021/jm950437v BindingDB Entry DOI: 10.7270/Q2CJ8CKR | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||