

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Serine--tRNA ligase (Escherichia coli (strain K12)) | BDBM50339906 (((2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihyd...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL MCE MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | n/a | 1.30 | n/a | n/a | n/a | n/a | n/a |
University of Warwick Curated by ChEMBL | Assay Description Binding affinity to C-terminal His10-tagged Escherichia coli B ER2560 SerRS expressed in Lemo21(DE3) SerRS assessed as dissociation constant by isoth... | J Med Chem 62: 9703-9717 (2019) Article DOI: 10.1021/acs.jmedchem.9b01131 BindingDB Entry DOI: 10.7270/Q2GX4G11 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Serine--tRNA ligase (Escherichia coli (strain K12)) | BDBM50339906 (((2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihyd...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL MCE MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | n/a | 1.30 | n/a | n/a | n/a | n/a | n/a |
University of Warwick Curated by ChEMBL | Assay Description Binding affinity to C-terminal His10-tagged Escherichia coli B ER2560 SerRS expressed in Lemo21(DE3) SerRS assessed as dissociation constant by isoth... | J Med Chem 62: 9703-9717 (2019) Article DOI: 10.1021/acs.jmedchem.9b01131 BindingDB Entry DOI: 10.7270/Q2GX4G11 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Serine--tRNA ligase (Escherichia coli (strain K12)) | BDBM50530859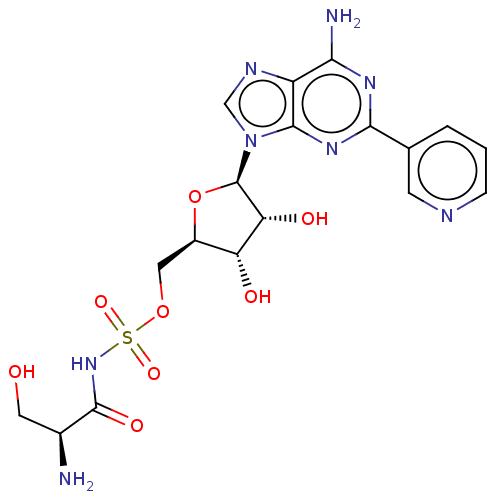 (CHEMBL4561900) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid PDB UniChem | PDB Article PubMed | n/a | n/a | n/a | 290 | n/a | n/a | n/a | n/a | n/a |
University of Warwick Curated by ChEMBL | Assay Description Binding affinity to C-terminal His10-tagged Escherichia coli B ER2560 SerRS expressed in Lemo21(DE3) SerRS assessed as affinity constant for first bi... | J Med Chem 62: 9703-9717 (2019) Article DOI: 10.1021/acs.jmedchem.9b01131 BindingDB Entry DOI: 10.7270/Q2GX4G11 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Serine--tRNA ligase (Escherichia coli (strain K12)) | BDBM50530859 (CHEMBL4561900) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid PDB UniChem | PDB Article PubMed | n/a | n/a | n/a | 290 | n/a | n/a | n/a | n/a | n/a |
University of Warwick Curated by ChEMBL | Assay Description Binding affinity to C-terminal His10-tagged Escherichia coli B ER2560 SerRS expressed in Lemo21(DE3) SerRS assessed as affinity constant for first bi... | J Med Chem 62: 9703-9717 (2019) Article DOI: 10.1021/acs.jmedchem.9b01131 BindingDB Entry DOI: 10.7270/Q2GX4G11 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Serine--tRNA ligase (Escherichia coli (strain K12)) | BDBM50530859 (CHEMBL4561900) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid PDB UniChem | PDB Article PubMed | n/a | n/a | n/a | 290 | n/a | n/a | n/a | n/a | n/a |
University of Warwick Curated by ChEMBL | Assay Description Binding affinity to C-terminal His10-tagged Escherichia coli B ER2560 SerRS expressed in Lemo21(DE3) SerRS assessed as dissociation constant by isoth... | J Med Chem 62: 9703-9717 (2019) Article DOI: 10.1021/acs.jmedchem.9b01131 BindingDB Entry DOI: 10.7270/Q2GX4G11 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Serine--tRNA ligase (Escherichia coli (strain K12)) | BDBM50530859 (CHEMBL4561900) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid PDB UniChem | PDB Article PubMed | n/a | n/a | n/a | 290 | n/a | n/a | n/a | n/a | n/a |
University of Warwick Curated by ChEMBL | Assay Description Binding affinity to C-terminal His10-tagged Escherichia coli B ER2560 SerRS expressed in Lemo21(DE3) SerRS assessed as dissociation constant by isoth... | J Med Chem 62: 9703-9717 (2019) Article DOI: 10.1021/acs.jmedchem.9b01131 BindingDB Entry DOI: 10.7270/Q2GX4G11 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Serine--tRNA ligase (Escherichia coli (strain K12)) | BDBM50530859 (CHEMBL4561900) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid PDB UniChem | PDB Article PubMed | n/a | n/a | n/a | 1.92E+3 | n/a | n/a | n/a | n/a | n/a |
University of Warwick Curated by ChEMBL | Assay Description Binding affinity to C-terminal His10-tagged Escherichia coli B ER2560 SerRS expressed in Lemo21(DE3) SerRS assessed as affinity constant for second b... | J Med Chem 62: 9703-9717 (2019) Article DOI: 10.1021/acs.jmedchem.9b01131 BindingDB Entry DOI: 10.7270/Q2GX4G11 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Serine--tRNA ligase (Escherichia coli (strain K12)) | BDBM50530859 (CHEMBL4561900) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid PDB UniChem | PDB Article PubMed | n/a | n/a | n/a | 1.92E+3 | n/a | n/a | n/a | n/a | n/a |
University of Warwick Curated by ChEMBL | Assay Description Binding affinity to C-terminal His10-tagged Escherichia coli B ER2560 SerRS expressed in Lemo21(DE3) SerRS assessed as affinity constant for second b... | J Med Chem 62: 9703-9717 (2019) Article DOI: 10.1021/acs.jmedchem.9b01131 BindingDB Entry DOI: 10.7270/Q2GX4G11 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||