

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Prothrombin (257/257 = 100%)† (Homo sapiens (Human)) | BDBM50075931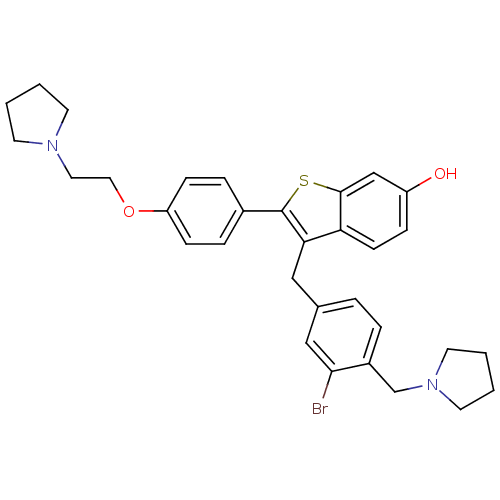 (3-(3-BROMO-4-PYRROLIDIN-1-YLMETHYL-BENZYL)-2-[4-PY...) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Patents Similars | PDB PubMed | 2.10 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Lilly Research Laboratories Curated by ChEMBL | Assay Description Binding inhibition against thrombin was measured by nonlinear regression analysis using Morrison's equation for tight-binding inhibition | Bioorg Med Chem Lett 9: 775-80 (1999) BindingDB Entry DOI: 10.7270/Q2PR7V46 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||