

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tyrosine-protein phosphatase non-receptor type 1 [1-298] (298/298 = 100%)† (Homo sapiens (Human)) | BDBM13958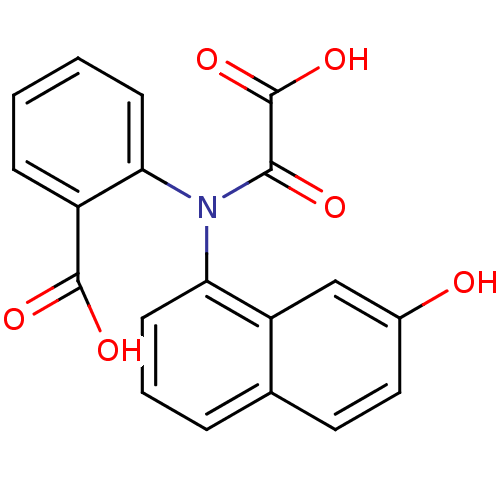 (2-[(7-Hydroxynaphthalen-1-yl)oxalylamino]benzoic A...) | PDB MMDB UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | DrugBank MMDB PC cid PC sid PDB UniChem Similars | DrugBank MMDB PDB Article PubMed | 1.70E+4 | -26.9 | n/a | 8.00E+3 | n/a | n/a | n/a | 7.5 | 22 |
Abbott Laboratories | Assay Description The phosphatase activity resulted in the formation of the colored product p-nitrophenol, which was continuously monitored at 405 nm every 30 s for 15... | J Med Chem 46: 2093-103 (2003) Article DOI: 10.1021/jm0205696 BindingDB Entry DOI: 10.7270/Q26H4FPH | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Tyrosine-protein phosphatase non-receptor type 1 (321/321 = 100%)† (Homo sapiens (Human)) | BDBM13958 (2-[(7-Hydroxynaphthalen-1-yl)oxalylamino]benzoic A...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | DrugBank MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | 2.51E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Abbott Laboratories Curated by ChEMBL | Assay Description Inhibition of PTP1B | J Med Chem 52: 3159-65 (2009) Article DOI: 10.1021/jm801444x BindingDB Entry DOI: 10.7270/Q2FF3TM8 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||