

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genome polyprotein (569/570 > 99%)† (Hepatitis C virus genotype 1b (isolate BK) (HCV)) | BDBM30545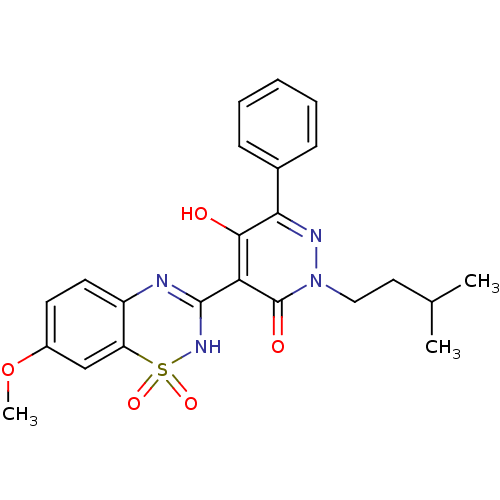 (pyridazinone derivative, 3) | PDB UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | DrugBank PC cid PC sid PDB UniChem Patents Similars | DrugBank PDB Article PubMed | n/a | n/a | 3.30E+4 | n/a | n/a | n/a | n/a | 7.5 | 28 |
Anadys Pharmaceuticals | Assay Description Assays were performed in a 96-well streptavidin-coated Flash-Plate using enzyme, RNA substrate, and [alpha-33P]GTP/GTP with inhibitor concentration v... | Bioorg Med Chem Lett 18: 1413-8 (2008) Article DOI: 10.1016/j.bmcl.2008.01.007 BindingDB Entry DOI: 10.7270/Q22N50MR | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||