

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kcal/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Serine protease 1 (223/223 = 100%)† (Bos taurus (bovine)) | BDBM12657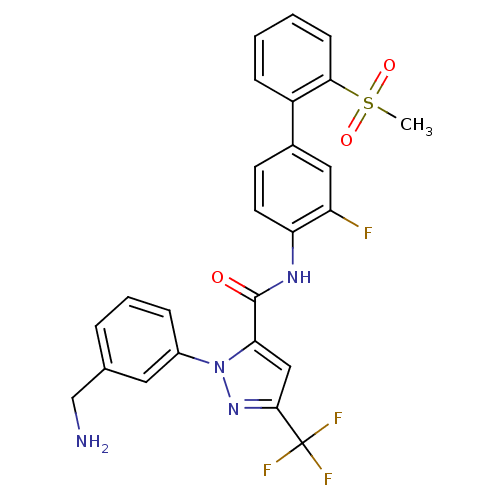 (1-[3-(Aminomethyl)phenyl]-N-[3-fluoro-2-(methylsul...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL MCE MMDB PC cid PC sid PDB UniChem Patents Similars | Article PubMed | 60 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Bristol-Myers Squibb Pharmaceutical Research Institute | Assay Description Ki values were obtained from human purified enzyme. All assays were run in microtiter plates. Plates were read for 30 min at 405 nm. Rates were deter... | J Med Chem 48: 1729-44 (2005) Article DOI: 10.1021/jm0497949 BindingDB Entry DOI: 10.7270/Q2BK19K9 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||