

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbonic anhydrase 2 (260/260 = 100%)† (Homo sapiens (Human)) | BDBM50380148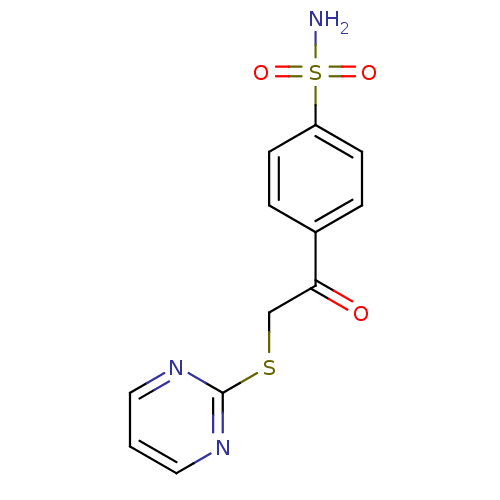 (CHEMBL2011156 | carbonic anhydrase (CA) inhibitors...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | n/a | n/a | n/a | n/a | 1.60E+6 | n/a | n/a |
Uppsala University Curated by ChEMBL | Assay Description Binding affinity to human recombinant CA2 expressed in Escherichia coli assessed as association rate constant after 30 secs by surface plasmon resona... | J Med Chem 59: 2083-93 (2016) Article DOI: 10.1021/acs.jmedchem.5b01723 BindingDB Entry DOI: 10.7270/Q2571DWK | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (docked) | ||||||||||||
| Carbonic anhydrase 2 (260/260 = 100%)† (Homo sapiens (Human)) | BDBM50380148 (CHEMBL2011156 | carbonic anhydrase (CA) inhibitors...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | n/a | n/a | n/a | 0.00800 | n/a | n/a | n/a |
Uppsala University Curated by ChEMBL | Assay Description Binding affinity to human recombinant CA2 expressed in Escherichia coli assessed as dissociation rate constant after 30 secs by surface plasmon reson... | J Med Chem 59: 2083-93 (2016) Article DOI: 10.1021/acs.jmedchem.5b01723 BindingDB Entry DOI: 10.7270/Q2571DWK | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (docked) | ||||||||||||
| Carbonic anhydrase 2 (260/260 = 100%)† (Homo sapiens (Human)) | BDBM50380148 (CHEMBL2011156 | carbonic anhydrase (CA) inhibitors...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | n/a | 5.20 | n/a | n/a | n/a | n/a | n/a |
Uppsala University Curated by ChEMBL | Assay Description Binding affinity to human recombinant CA2 expressed in Escherichia coli after 30 secs by surface plasmon resonance assay | J Med Chem 59: 2083-93 (2016) Article DOI: 10.1021/acs.jmedchem.5b01723 BindingDB Entry DOI: 10.7270/Q2571DWK | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (docked) | ||||||||||||
| Carbonic anhydrase 2 (260/260 = 100%)† (Homo sapiens (Human)) | BDBM50380148 (CHEMBL2011156 | carbonic anhydrase (CA) inhibitors...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | n/a | 7.10 | n/a | n/a | n/a | n/a | n/a |
Uppsala University Curated by ChEMBL | Assay Description Binding affinity to human recombinant CA2 expressed in Escherichia coli by fluorescence-based thermal shift assay | J Med Chem 59: 2083-93 (2016) Article DOI: 10.1021/acs.jmedchem.5b01723 BindingDB Entry DOI: 10.7270/Q2571DWK | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (docked) | ||||||||||||
| Carbonic anhydrase 2 (260/260 = 100%)† (Homo sapiens (Human)) | BDBM50380148 (CHEMBL2011156 | carbonic anhydrase (CA) inhibitors...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | n/a | 17 | n/a | n/a | n/a | n/a | n/a |
Vilnius University Curated by ChEMBL | Assay Description Binding affinity to human recombinant carbonic anhydrase 2 by thermal shift assay | Bioorg Med Chem 21: 6937-47 (2013) Article DOI: 10.1016/j.bmc.2013.09.029 BindingDB Entry DOI: 10.7270/Q2S46VZ9 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (docked) | ||||||||||||
| Carbonic anhydrase 2 (260/260 = 100%)† (Homo sapiens (Human)) | BDBM50380148 (CHEMBL2011156 | carbonic anhydrase (CA) inhibitors...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | n/a | 17 | n/a | n/a | n/a | n/a | n/a |
Vilnius University Curated by ChEMBL | Assay Description Binding affinity to human recombinant carbonic anhydrase-2 by thermal shift assay | Eur J Med Chem 51: 259-70 (2012) Article DOI: 10.1016/j.ejmech.2012.02.050 BindingDB Entry DOI: 10.7270/Q2FJ2HSF | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (docked) | ||||||||||||
| Carbonic anhydrase 2 (260/260 = 100%)† (Homo sapiens (Human)) | BDBM50380148 (CHEMBL2011156 | carbonic anhydrase (CA) inhibitors...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | n/a | n/a | n/a | 0.00800 | n/a | n/a | n/a |
Vilnius University Curated by ChEMBL | Assay Description Binding affinity to human full length His-tagged CA2 (1 to 260 residues) expressed in Escherichia coli BL21(DE3) assessed as dissociation rate consta... | J Med Chem 61: 2292-2302 (2018) Article DOI: 10.1021/acs.jmedchem.7b01408 BindingDB Entry DOI: 10.7270/Q23T9KNS | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (docked) | ||||||||||||
| Carbonic anhydrase 2 (260/260 = 100%)† (Homo sapiens (Human)) | BDBM50380148 (CHEMBL2011156 | carbonic anhydrase (CA) inhibitors...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | n/a | 0.0170 | n/a | n/a | n/a | n/a | n/a |
Vilnius University Curated by ChEMBL | Assay Description Binding affinity to human full length His-tagged CA2 (1 to 260 residues) expressed in Escherichia coli BL21(DE3) assessed as intrinsic thermodynamic ... | J Med Chem 61: 2292-2302 (2018) Article DOI: 10.1021/acs.jmedchem.7b01408 BindingDB Entry DOI: 10.7270/Q23T9KNS | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (docked) | ||||||||||||
| Carbonic anhydrase 2 (260/260 = 100%)† (Homo sapiens (Human)) | BDBM50380148 (CHEMBL2011156 | carbonic anhydrase (CA) inhibitors...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | n/a | n/a | n/a | 12 | n/a | n/a | n/a | n/a | n/a |
Vilnius University Curated by ChEMBL | Assay Description Binding affinity to human recombinant carbonic anhydrase-2 by isothermal titration calorimetry assay | Eur J Med Chem 51: 259-70 (2012) Article DOI: 10.1016/j.ejmech.2012.02.050 BindingDB Entry DOI: 10.7270/Q2FJ2HSF | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (docked) | ||||||||||||