

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cellular tumor antigen p53 [1-83]/E3 ubiquitin-protein ligase Mdm2 [1-188] (95/95 = 100%)† (Homo sapiens (Human)) | BDBM50388626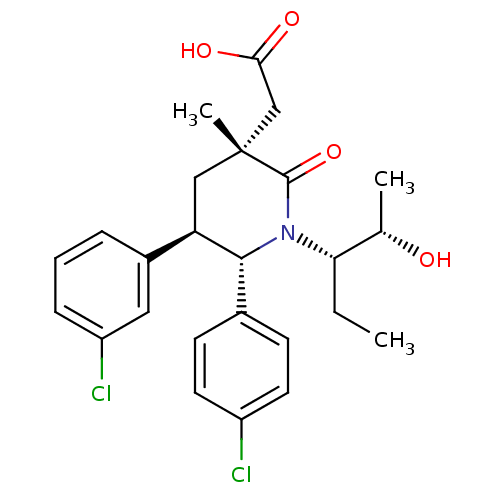 (CHEMBL2059435 | US9296736, 152 | US9593129, Exampl...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL MCE MMDB PC cid PC sid PDB UniChem Similars | PDB US Patent | n/a | n/a | 1 | n/a | n/a | n/a | n/a | 7.4 | 25 |
Amgen INC. US Patent | Assay Description The standard assay conditions for the in vitro HTRF assay consisted of a 50 ul total reaction volume in black 384-well Costar polypropylene plates in... | US Patent US9296736 (2016) BindingDB Entry DOI: 10.7270/Q29022M2 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| E3 ubiquitin-protein ligase Mdm2 [1-188] (95/95 = 100%)† (Homo sapiens (Human)) | BDBM50388626 (CHEMBL2059435 | US9296736, 152 | US9593129, Exampl...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL MCE MMDB PC cid PC sid PDB UniChem Similars | PDB US Patent | n/a | n/a | 1 | n/a | n/a | n/a | n/a | n/a | n/a |
Amgen, Inc. US Patent | Assay Description The standard assay conditions for the in vitro HTRF assay consisted of a 50 ul total reaction volume in black 384-well Costar polypropylene plates in... | US Patent US9593129 (2017) BindingDB Entry DOI: 10.7270/Q20P1232 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| E3 ubiquitin-protein ligase Mdm2 (95/95 = 100%)† (Homo sapiens (Human)) | BDBM50388626 (CHEMBL2059435 | US9296736, 152 | US9593129, Exampl...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MCE MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | 1.10 | n/a | n/a | n/a | n/a | n/a | n/a |
Amgen Inc. Curated by ChEMBL | Assay Description Binding affinity to human GST-thrombin-tagged MDM2 assessed as inhibition of interaction with human p53 after 1 hr by HTRF assay | J Med Chem 57: 1454-72 (2014) Article DOI: 10.1021/jm401753e BindingDB Entry DOI: 10.7270/Q24M960Z | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| E3 ubiquitin-protein ligase Mdm2 (95/95 = 100%)† (Homo sapiens (Human)) | BDBM50388626 (CHEMBL2059435 | US9296736, 152 | US9593129, Exampl...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MCE MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | 4.10 | n/a | n/a | n/a | n/a | n/a | n/a |
Amgen Inc. Curated by ChEMBL | Assay Description Binding affinity to human GST-thrombin-tagged MDM2 assessed as inhibition of interaction with human p53 after 1 hr by HTRF assay in presence of 15% h... | J Med Chem 57: 1454-72 (2014) Article DOI: 10.1021/jm401753e BindingDB Entry DOI: 10.7270/Q24M960Z | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Cellular tumor antigen p53 [1-83]/E3 ubiquitin-protein ligase Mdm2 [1-188] (95/95 = 100%)† (Homo sapiens (Human)) | BDBM50388626 (CHEMBL2059435 | US9296736, 152 | US9593129, Exampl...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL MCE MMDB PC cid PC sid PDB UniChem Similars | PDB US Patent | n/a | n/a | 10 | n/a | n/a | n/a | n/a | 7.4 | 25 |
Amgen INC. US Patent | Assay Description The standard assay conditions for the in vitro HTRF assay consisted of a 50 ul total reaction volume in black 384-well Costar polypropylene plates in... | US Patent US9296736 (2016) BindingDB Entry DOI: 10.7270/Q29022M2 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| E3 ubiquitin-protein ligase Mdm2 [1-188] (95/95 = 100%)† (Homo sapiens (Human)) | BDBM50388626 (CHEMBL2059435 | US9296736, 152 | US9593129, Exampl...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL MCE MMDB PC cid PC sid PDB UniChem Similars | PDB US Patent | n/a | n/a | 10 | n/a | n/a | n/a | n/a | n/a | n/a |
Amgen, Inc. US Patent | Assay Description The standard assay conditions for the in vitro HTRF assay consisted of a 50 ul total reaction volume in black 384-well Costar polypropylene plates in... | US Patent US9593129 (2017) BindingDB Entry DOI: 10.7270/Q20P1232 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| E3 ubiquitin-protein ligase Mdm2 (95/95 = 100%)† (Homo sapiens (Human)) | BDBM50388626 (CHEMBL2059435 | US9296736, 152 | US9593129, Exampl...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MCE MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | n/a | 0.400 | n/a | n/a | n/a | n/a | n/a |
Amgen Inc. Curated by ChEMBL | Assay Description Binding affinity to human MDM2 by SPR analysis | J Med Chem 55: 4936-54 (2012) Article DOI: 10.1021/jm300354j BindingDB Entry DOI: 10.7270/Q2S75HGJ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| E3 ubiquitin-protein ligase Mdm2 (95/95 = 100%)† (Homo sapiens (Human)) | BDBM50388626 (CHEMBL2059435 | US9296736, 152 | US9593129, Exampl...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MCE MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | n/a | 0.400 | n/a | n/a | n/a | n/a | n/a |
University of Michigan Comprehensive Cancer Center Curated by ChEMBL | Assay Description Binding affinity to human MDM2 by surface plasmon resonance binding assay | J Med Chem 55: 4934-5 (2012) Article DOI: 10.1021/jm3007068 BindingDB Entry DOI: 10.7270/Q2HT2QCX | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| E3 ubiquitin-protein ligase Mdm2 (95/95 = 100%)† (Homo sapiens (Human)) | BDBM50388626 (CHEMBL2059435 | US9296736, 152 | US9593129, Exampl...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL MCE MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | n/a | 0.400 | n/a | n/a | n/a | n/a | n/a |
Amgen Inc. Curated by ChEMBL | Assay Description Binding affinity to human MDM2 by by Surface Plasmon Resonace (SPR) spectroscopy binding assay | J Med Chem 57: 1454-72 (2014) Article DOI: 10.1021/jm401753e BindingDB Entry DOI: 10.7270/Q24M960Z | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||