 Found 6 hits Enzyme Inhibition Constant Data
Found 6 hits Enzyme Inhibition Constant Data Target/Host
(Institution) | Ligand | Target/Host
Links | Ligand
Links | Trg + Lig
Links | Ki
nM | ΔG°
kJ/mole | IC50
nM | Kd
nM | EC50/IC50
nM | koff
s-1 | kon
M-1s-1 | pH | Temp
°C |
|---|
DNA polymerase alpha catalytic subunit
(921/922 > 99%)†
(Homo sapiens (Human)) | BDBM50090910
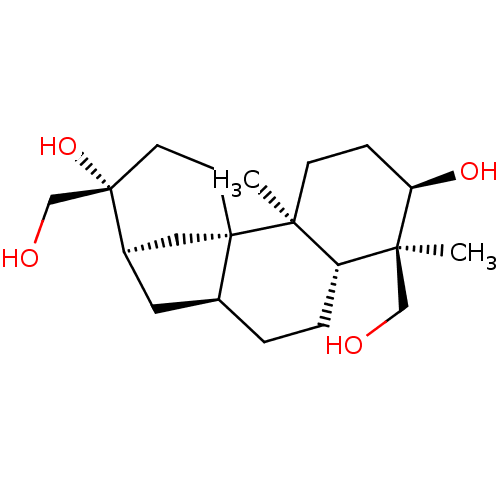
(3alpha,16,17,18-tetrahydroxyaphidicolone | 6,13-di...)Show SMILES C[C@@]1(CO)[C@H](O)CC[C@@]2(C)[C@H]1CC[C@H]1C[C@@H]3C[C@]21CC[C@]3(O)CO |r| Show InChI InChI=1S/C20H34O4/c1-17(11-21)15-4-3-13-9-14-10-19(13,7-8-20(14,24)12-22)18(15,2)6-5-16(17)23/h13-16,21-24H,3-12H2,1-2H3/t13-,14+,15-,16+,17-,18-,19-,20-/m0/s1 | PDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| n/a | n/a | 2.40E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
Emory University School of Medicine
Curated by ChEMBL
| Assay Description
Inhibition of human DNA polymerase alpha using 5' end radiolabeled 24nt DNA/48nt DNA as primer/template after 5 mins by PAGE analysis |
J Med Chem 60: 5424-5437 (2017)
Article DOI: 10.1021/acs.jmedchem.7b00067
BindingDB Entry DOI: 10.7270/Q2BZ6873 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
DNA polymerase alpha catalytic subunit
(921/922 > 99%)†
(Homo sapiens (Human)) | BDBM50090910

(3alpha,16,17,18-tetrahydroxyaphidicolone | 6,13-di...)Show SMILES C[C@@]1(CO)[C@H](O)CC[C@@]2(C)[C@H]1CC[C@H]1C[C@@H]3C[C@]21CC[C@]3(O)CO |r| Show InChI InChI=1S/C20H34O4/c1-17(11-21)15-4-3-13-9-14-10-19(13,7-8-20(14,24)12-22)18(15,2)6-5-16(17)23/h13-16,21-24H,3-12H2,1-2H3/t13-,14+,15-,16+,17-,18-,19-,20-/m0/s1 | PDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| n/a | n/a | 2.60E+3 | n/a | n/a | n/a | n/a | 7.5 | n/a |
Pharmacia Corporation
Curated by ChEMBL
| Assay Description
Inhibition of human alpha DNA polymerase (95 uL) activity in a solution containg 6.4 mM HEPES (pH 7.5) upon incubation for 12 minutes at 26 degrees C... |
J Med Chem 48: 5794-804 (2005)
Article DOI: 10.1021/jm050162b
BindingDB Entry DOI: 10.7270/Q2G44R39 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
DNA polymerase alpha catalytic subunit
(921/922 > 99%)†
(Homo sapiens (Human)) | BDBM50090910

(3alpha,16,17,18-tetrahydroxyaphidicolone | 6,13-di...)Show SMILES C[C@@]1(CO)[C@H](O)CC[C@@]2(C)[C@H]1CC[C@H]1C[C@@H]3C[C@]21CC[C@]3(O)CO |r| Show InChI InChI=1S/C20H34O4/c1-17(11-21)15-4-3-13-9-14-10-19(13,7-8-20(14,24)12-22)18(15,2)6-5-16(17)23/h13-16,21-24H,3-12H2,1-2H3/t13-,14+,15-,16+,17-,18-,19-,20-/m0/s1 | PDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| n/a | n/a | 2.60E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
Pfizer, Inc.
Curated by ChEMBL
| Assay Description
Inhibition of human DNA polymerase alpha |
Bioorg Med Chem Lett 20: 3039-42 (2010)
Article DOI: 10.1016/j.bmcl.2010.03.115
BindingDB Entry DOI: 10.7270/Q2000281 |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
DNA polymerase alpha catalytic subunit
(921/922 > 99%)†
(Homo sapiens (Human)) | BDBM50090910

(3alpha,16,17,18-tetrahydroxyaphidicolone | 6,13-di...)Show SMILES C[C@@]1(CO)[C@H](O)CC[C@@]2(C)[C@H]1CC[C@H]1C[C@@H]3C[C@]21CC[C@]3(O)CO |r| Show InChI InChI=1S/C20H34O4/c1-17(11-21)15-4-3-13-9-14-10-19(13,7-8-20(14,24)12-22)18(15,2)6-5-16(17)23/h13-16,21-24H,3-12H2,1-2H3/t13-,14+,15-,16+,17-,18-,19-,20-/m0/s1 | PDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
PubMed
| n/a | n/a | 3.00E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
Pharmacia
Curated by ChEMBL
| Assay Description
Compound was evaluated for the inhibitory activity against Human alpha polymerase |
Bioorg Med Chem Lett 10: 2079-81 (2000)
BindingDB Entry DOI: 10.7270/Q2222T0M |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
DNA polymerase alpha catalytic subunit
(921/922 > 99%)†
(Homo sapiens (Human)) | BDBM50090910

(3alpha,16,17,18-tetrahydroxyaphidicolone | 6,13-di...)Show SMILES C[C@@]1(CO)[C@H](O)CC[C@@]2(C)[C@H]1CC[C@H]1C[C@@H]3C[C@]21CC[C@]3(O)CO |r| Show InChI InChI=1S/C20H34O4/c1-17(11-21)15-4-3-13-9-14-10-19(13,7-8-20(14,24)12-22)18(15,2)6-5-16(17)23/h13-16,21-24H,3-12H2,1-2H3/t13-,14+,15-,16+,17-,18-,19-,20-/m0/s1 | PDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
Article
PubMed
| n/a | n/a | 1.60E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
303A College Road East
Curated by ChEMBL
| Assay Description
Inhibition of human DNA polymerase alpha assessed as incorporation of [alpha-32P]dCTP up to 1mM preincubated for 30 mins by whatman DE81 paper bindin... |
Antimicrob Agents Chemother 54: 3187-96 (2010)
Article DOI: 10.1128/AAC.00399-10
BindingDB Entry DOI: 10.7270/Q2G1614S |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |
DNA polymerase alpha catalytic subunit
(921/922 > 99%)†
(Homo sapiens (Human)) | BDBM50090910

(3alpha,16,17,18-tetrahydroxyaphidicolone | 6,13-di...)Show SMILES C[C@@]1(CO)[C@H](O)CC[C@@]2(C)[C@H]1CC[C@H]1C[C@@H]3C[C@]21CC[C@]3(O)CO |r| Show InChI InChI=1S/C20H34O4/c1-17(11-21)15-4-3-13-9-14-10-19(13,7-8-20(14,24)12-22)18(15,2)6-5-16(17)23/h13-16,21-24H,3-12H2,1-2H3/t13-,14+,15-,16+,17-,18-,19-,20-/m0/s1 | PDB
NCI pathway
Reactome pathway
KEGG
UniProtKB/SwissProt
DrugBank
antibodypedia
GoogleScholar
AffyNet 
| Purchase
CHEMBL
MCE
PC cid
PC sid
PDB
UniChem
Patents
Similars
| PDB
PubMed
| n/a | n/a | n/a | n/a | 2.00E+3 | n/a | n/a | n/a | n/a |
Wellcome Research Laboratories
Curated by ChEMBL
| Assay Description
Effective concentration against DNA polymerase alpha |
J Med Chem 36: 3503-10 (1994)
BindingDB Entry DOI: 10.7270/Q2HT2PXP |
More data for this
Ligand-Target Pair | 
3D Structure (crystal) |


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data