

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glutamate receptor ionotropic, NMDA 1 (738/816 = 90%)† (RAT) | BDBM50344263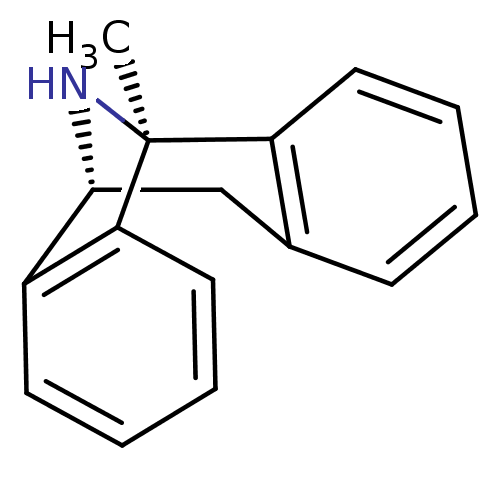 ((+)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]he...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL MCE KEGG PC cid PC sid PDB UniChem Patents Similars | PDB PubMed | 9 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
National Institute of Diabetes and Digestive and Kidney Diseases Curated by ChEMBL | Assay Description Ability to displace [3H]TCP from high affinity PCP N-methyl-D-aspartate glutamate receptor in rat brain homogenatesin vitro was determined | J Med Chem 33: 1069-76 (1990) BindingDB Entry DOI: 10.7270/Q2J38T4C | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Glutamate receptor ionotropic, NMDA 1 (738/816 = 90%)† (RAT) | BDBM50344263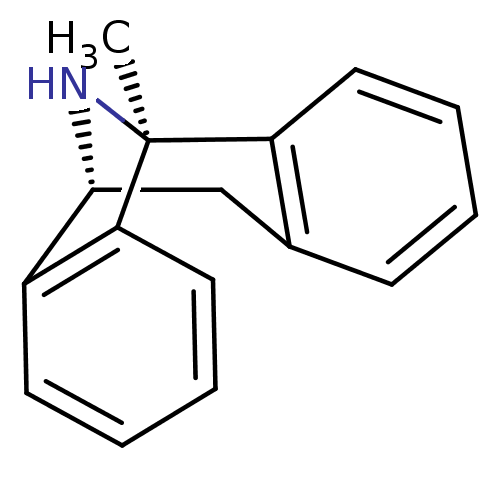 ((+)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]he...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL MCE KEGG PC cid PC sid PDB UniChem Patents Similars | PDB PubMed | 31 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
National Institute of Diabetes and Digestive and Kidney Diseases Curated by ChEMBL | Assay Description Ability to displace [3H]TCP from high affinity PCP N-methyl-D-aspartate glutamate receptor in rat brain homogenates in vitro was determined | J Med Chem 33: 1069-76 (1990) BindingDB Entry DOI: 10.7270/Q2J38T4C | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Glutamate receptor ionotropic, NMDA 1 (738/816 = 90%)† (RAT) | BDBM50344263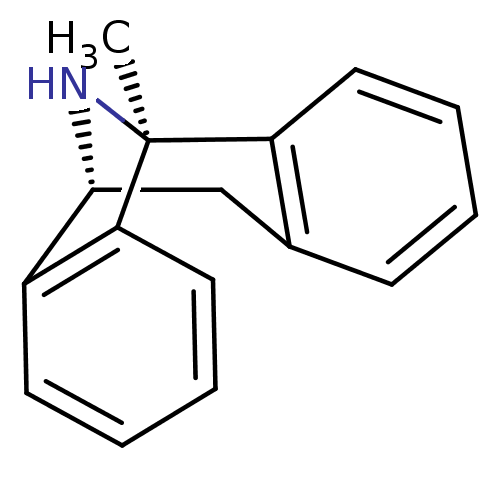 ((+)-1-methyl-16-azatetracyclo[7.6.1.02,7.010,15]he...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL MCE KEGG PC cid PC sid PDB UniChem Patents Similars | PDB PubMed | 31 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
National Institute of Diabetes and Digestive and Kidney Diseases Curated by ChEMBL | Assay Description Ability to displace [3H]TCP from high affinity PCP N-methyl-D-aspartate glutamate receptor in rat brain homogenatesin vitro was determined | J Med Chem 33: 1069-76 (1990) BindingDB Entry DOI: 10.7270/Q2J38T4C | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Glutamate receptor ionotropic, NMDA 2B (733/845 = 87%)† (Rattus norvegicus (Rat)) | BDBM50030386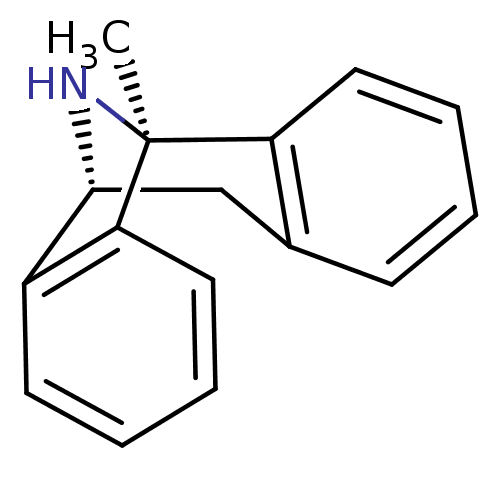 ((1S)-1-methyl-16-azatetracyclo[7.6.1.0^{2,7}.0^{10...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | Purchase MCE KEGG PC cid PC sid PDB UniChem Similars | PDB Article PubMed | >1.00E+5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Istituto Italiano di Tecnologia Curated by ChEMBL | Assay Description Displacement of [3H]Ifenprodil from NMDAR-2B in Sprague-Dawley rat frontal cortex homogenates after 2 hrs by liquid scintillation counting | J Med Chem 55: 9708-21 (2012) Article DOI: 10.1021/jm3009458 BindingDB Entry DOI: 10.7270/Q2VQ33TR | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||