

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tyrosine-protein phosphatase non-receptor type 1 [1-298] (Homo sapiens (Human)) | BDBM13809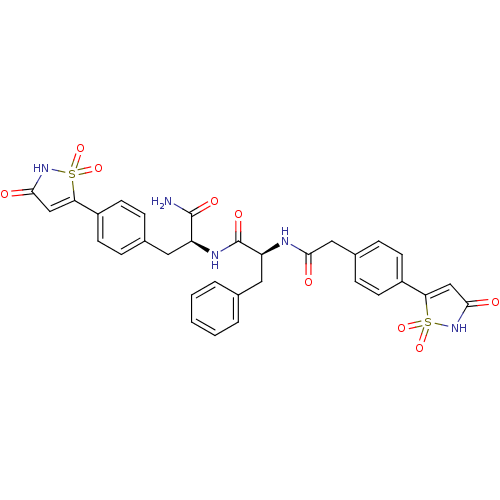 ((2S)-N-[(1S)-1-carbamoyl-2-[4-(1,1,3-trioxo-2,3-di...) | PDB MMDB UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | n/a | n/a | 65 | n/a | n/a | n/a | n/a | 7.0 | 22 |
Incyte Corporation | Assay Description The activity of PTP1B enzyme was assayed with 4-nitrophenyl phosphate (pNPP) as substrate. Rate of formation of the phenolate ion was monitored at 41... | J Biol Chem 281: 32784-95 (2006) Article DOI: 10.1074/jbc.M606873200 BindingDB Entry DOI: 10.7270/Q2GX48SD | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Tyrosine-protein phosphatase non-receptor type 1 [1-298] (Homo sapiens (Human)) | BDBM13808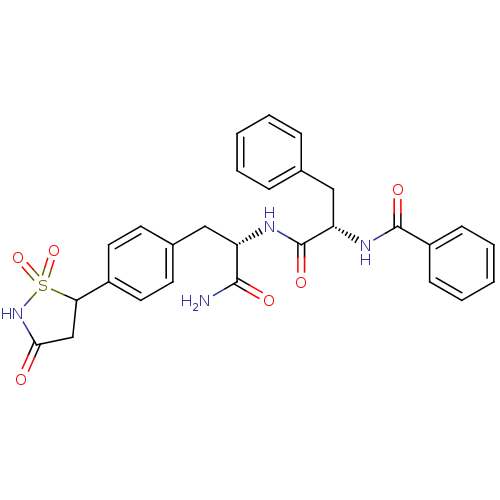 ((2S)-N-[(1S)-1-carbamoyl-2-[4-(1,1,3-trioxo-1,2-th...) | PDB MMDB UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | n/a | n/a | 185 | n/a | n/a | n/a | n/a | 7.0 | 22 |
Incyte Corporation | Assay Description The activity of PTP1B enzyme was assayed with 4-nitrophenyl phosphate (pNPP) as substrate. Rate of formation of the phenolate ion was monitored at 41... | J Biol Chem 281: 32784-95 (2006) Article DOI: 10.1074/jbc.M606873200 BindingDB Entry DOI: 10.7270/Q2GX48SD | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Tyrosine-protein phosphatase non-receptor type 1 [1-298] (Homo sapiens (Human)) | BDBM13465 ((2S)-N-[(1S)-1-carbamoyl-2-{4-[(5S)-1,1,3-trioxo-1...) | PDB MMDB UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL DrugBank MMDB PC cid PC sid PDB UniChem Similars | DrugBank MMDB PDB Article PubMed | n/a | n/a | 190 | n/a | n/a | n/a | n/a | 7.0 | 22 |
Incyte Corporation | Assay Description The activity of PTP1B enzyme was assayed with 4-nitrophenyl phosphate (pNPP) as substrate. Rate of formation of the phenolate ion was monitored at 41... | J Biol Chem 281: 32784-95 (2006) Article DOI: 10.1074/jbc.M606873200 BindingDB Entry DOI: 10.7270/Q2GX48SD | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Tyrosine-protein phosphatase non-receptor type 1 [1-298] (Homo sapiens (Human)) | BDBM13807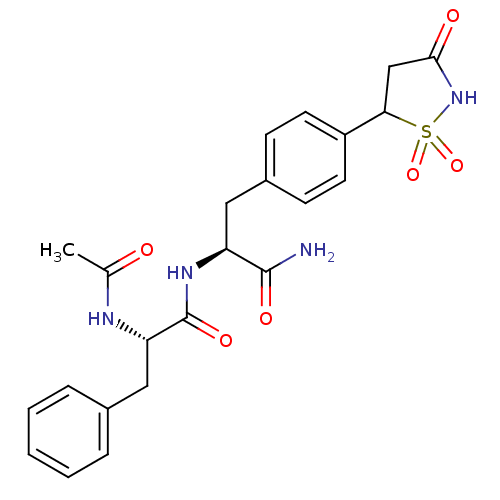 ((2S)-N-[(1S)-1-carbamoyl-2-[4-(1,1,3-trioxo-1,2-th...) | PDB MMDB UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | n/a | n/a | 210 | n/a | n/a | n/a | n/a | 7.0 | 22 |
Incyte Corporation | Assay Description The activity of PTP1B enzyme was assayed with 4-nitrophenyl phosphate (pNPP) as substrate. Rate of formation of the phenolate ion was monitored at 41... | J Biol Chem 281: 32784-95 (2006) Article DOI: 10.1074/jbc.M606873200 BindingDB Entry DOI: 10.7270/Q2GX48SD | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Tyrosine-protein phosphatase non-receptor type 1 [1-298] (Homo sapiens (Human)) | BDBM13806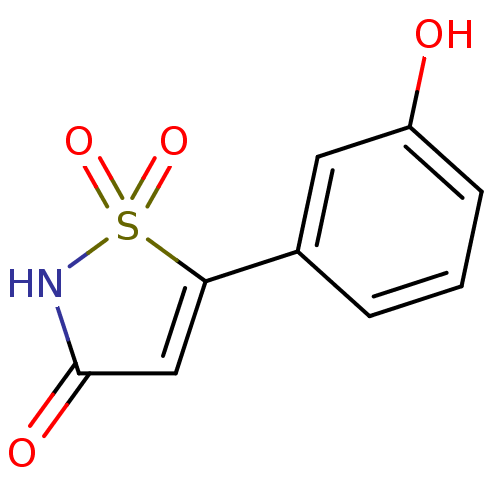 (5-(3-hydroxyphenyl)-2,3-dihydro-1,2-thiazole-1,1,3...) | PDB MMDB UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | DrugBank PC cid PC sid PDB UniChem Patents | DrugBank PDB Article PubMed | n/a | n/a | 1.35E+3 | n/a | n/a | n/a | n/a | 7.0 | 22 |
Incyte Corporation | Assay Description The activity of PTP1B enzyme was assayed with 4-nitrophenyl phosphate (pNPP) as substrate. Rate of formation of the phenolate ion was monitored at 41... | J Biol Chem 281: 32784-95 (2006) Article DOI: 10.1074/jbc.M606873200 BindingDB Entry DOI: 10.7270/Q2GX48SD | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Tyrosine-protein phosphatase non-receptor type 1 [1-298] (Homo sapiens (Human)) | BDBM13472 (({4-[(2S)-2-carbamoyl-2-[(2S)-2-acetamido-3-phenyl...) | PDB MMDB UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL DrugBank MMDB PC cid PC sid PDB UniChem Similars | DrugBank MMDB PDB Article PubMed | n/a | n/a | 1.75E+3 | n/a | n/a | n/a | n/a | 7.0 | 22 |
Incyte Corporation | Assay Description The activity of PTP1B enzyme was assayed with 4-nitrophenyl phosphate (pNPP) as substrate. Rate of formation of the phenolate ion was monitored at 41... | J Biol Chem 281: 32784-95 (2006) Article DOI: 10.1074/jbc.M606873200 BindingDB Entry DOI: 10.7270/Q2GX48SD | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Tyrosine-protein phosphatase non-receptor type 1 [1-298] (Homo sapiens (Human)) | BDBM13811 ((2S)-N-[(1S)-1-carbamoyl-2-[4-(1,1,3-trioxo-2,3-di...) | PDB MMDB UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | Article PubMed | n/a | n/a | 3.00E+3 | n/a | n/a | n/a | n/a | 7.0 | 22 |
Incyte Corporation | Assay Description The activity of PTP1B enzyme was assayed with 4-nitrophenyl phosphate (pNPP) as substrate. Rate of formation of the phenolate ion was monitored at 41... | J Biol Chem 281: 32784-95 (2006) Article DOI: 10.1074/jbc.M606873200 BindingDB Entry DOI: 10.7270/Q2GX48SD | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (docked) | ||||||||||||
| Tyrosine-protein phosphatase non-receptor type 1 [1-298] (Homo sapiens (Human)) | BDBM13812 ((2S)-N-[(1S)-1-carbamoyl-2-{4-[(5R)-1,1,3-trioxo-1...) | PDB MMDB UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 1.60E+4 | n/a | n/a | n/a | n/a | 7.0 | 22 |
Incyte Corporation | Assay Description The activity of PTP1B enzyme was assayed with 4-nitrophenyl phosphate (pNPP) as substrate. Rate of formation of the phenolate ion was monitored at 41... | J Biol Chem 281: 32784-95 (2006) Article DOI: 10.1074/jbc.M606873200 BindingDB Entry DOI: 10.7270/Q2GX48SD | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (docked) | ||||||||||||