

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glutamate receptor ionotropic, NMDA 1/2C (RAT-Rattus norvegicus (Rat)) | BDBM50134218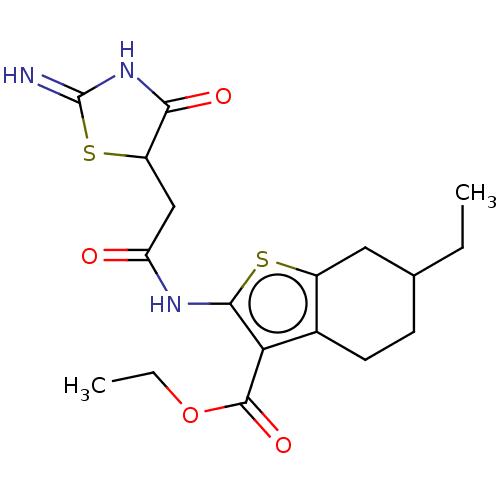 (CHEMBL3741652) | PDB KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | Article PubMed | n/a | n/a | 900 | n/a | n/a | n/a | n/a | n/a | n/a |
Emory University Curated by ChEMBL | Assay Description Negative allosteric modulation of rat GluN1a/GluN2C receptor expressed in Xenopus laevis oocytes assessed as suppression of 100 uM glutamate and 30 u... | Bioorg Med Chem Lett 25: 5583-8 (2015) Article DOI: 10.1016/j.bmcl.2015.10.046 BindingDB Entry DOI: 10.7270/Q2BK1F61 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Glutamate receptor ionotropic, NMDA 1/2A (Rattus norvegicus (Rat)-RAT) | BDBM50135179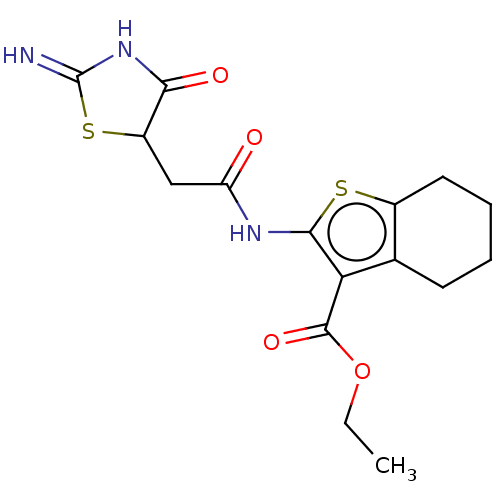 (CHEMBL568556) | PDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 2.30E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
Emory University Curated by ChEMBL | Assay Description Negative allosteric modulation of rat GluN1a/GluN2A receptor expressed in Xenopus laevis oocytes assessed as suppression of 100 uM glutamate and 30 u... | Bioorg Med Chem Lett 25: 5583-8 (2015) Article DOI: 10.1016/j.bmcl.2015.10.046 BindingDB Entry DOI: 10.7270/Q2BK1F61 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Glutamate receptor ionotropic, NMDA 1/2A (Rattus norvegicus (Rat)-RAT) | BDBM50134214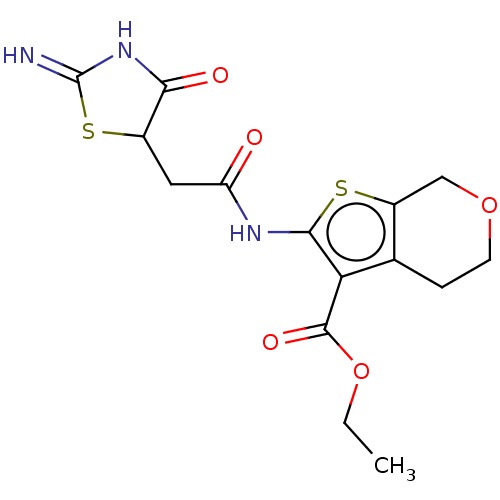 (CHEMBL3740292) | PDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents | Article PubMed | n/a | n/a | 3.00E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
Emory University Curated by ChEMBL | Assay Description Negative allosteric modulation of rat GluN1a/GluN2A receptor expressed in Xenopus laevis oocytes assessed as suppression of 100 uM glutamate and 30 u... | Bioorg Med Chem Lett 25: 5583-8 (2015) Article DOI: 10.1016/j.bmcl.2015.10.046 BindingDB Entry DOI: 10.7270/Q2BK1F61 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Glutamate receptor ionotropic, NMDA 1/2A (Rattus norvegicus (Rat)-RAT) | BDBM50134216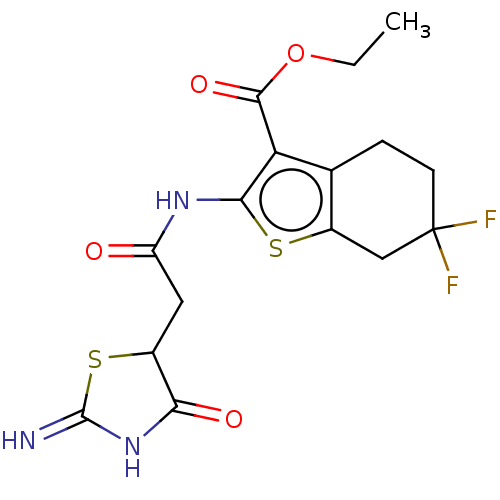 (CHEMBL3740010) | PDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | Article PubMed | n/a | n/a | 5.20E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
Emory University Curated by ChEMBL | Assay Description Negative allosteric modulation of rat GluN1a/GluN2A receptor expressed in Xenopus laevis oocytes assessed as suppression of 100 uM glutamate and 30 u... | Bioorg Med Chem Lett 25: 5583-8 (2015) Article DOI: 10.1016/j.bmcl.2015.10.046 BindingDB Entry DOI: 10.7270/Q2BK1F61 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Glutamate receptor ionotropic, NMDA 1/2A (Rattus norvegicus (Rat)-RAT) | BDBM50134215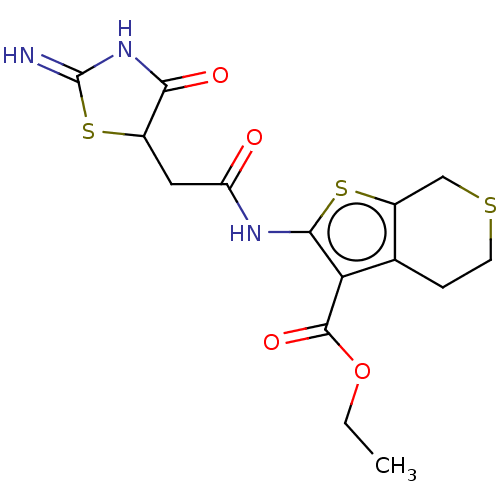 (CHEMBL3740638) | PDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents | Article PubMed | n/a | n/a | 6.20E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
Emory University Curated by ChEMBL | Assay Description Negative allosteric modulation of rat GluN1a/GluN2A receptor expressed in Xenopus laevis oocytes assessed as suppression of 100 uM glutamate and 30 u... | Bioorg Med Chem Lett 25: 5583-8 (2015) Article DOI: 10.1016/j.bmcl.2015.10.046 BindingDB Entry DOI: 10.7270/Q2BK1F61 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Glutamate receptor ionotropic, NMDA 1/2A (Rattus norvegicus (Rat)-RAT) | BDBM50134217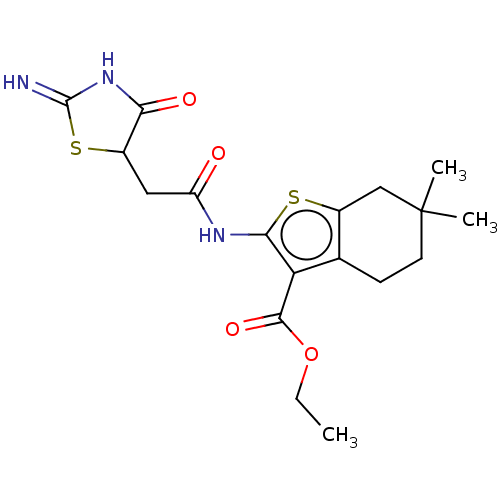 (CHEMBL3740320) | PDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 1.61E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
Emory University Curated by ChEMBL | Assay Description Negative allosteric modulation of rat GluN1a/GluN2A receptor expressed in Xenopus laevis oocytes assessed as suppression of 100 uM glutamate and 30 u... | Bioorg Med Chem Lett 25: 5583-8 (2015) Article DOI: 10.1016/j.bmcl.2015.10.046 BindingDB Entry DOI: 10.7270/Q2BK1F61 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Glutamate receptor ionotropic, NMDA 1/2B (Rattus norvegicus (Rat)-RAT) | BDBM50135181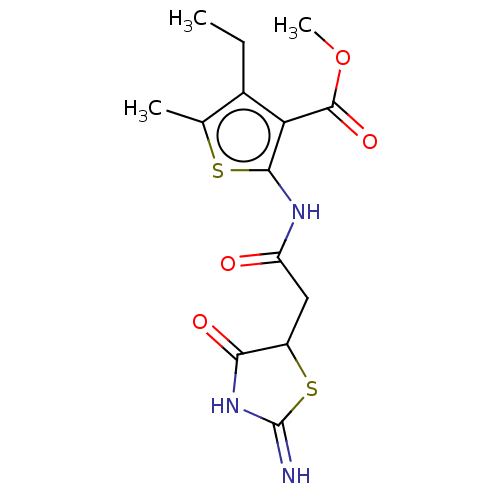 (CHEMBL3740575) | PDB KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL PC cid PC sid UniChem | Article PubMed | n/a | n/a | 4.38E+4 | n/a | n/a | n/a | n/a | n/a | n/a |
Emory University Curated by ChEMBL | Assay Description Negative allosteric modulation of rat GluN1a/GluN2B receptor expressed in Xenopus laevis oocytes assessed as suppression of 100 uM glutamate and 30 u... | Bioorg Med Chem Lett 25: 5583-8 (2015) Article DOI: 10.1016/j.bmcl.2015.10.046 BindingDB Entry DOI: 10.7270/Q2BK1F61 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||