

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ATP-dependent Clp protease proteolytic subunit (Escherichia coli (strain K12)) | BDBM50535810 (CHEMBL4593796) | PDB KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | n/a | 2.54E+4 | n/a | n/a | n/a | n/a |
University of Oklahoma COBRE in Structural Biology Curated by ChEMBL | Assay Description Activation of Escherichia coli BLR (DE3) ClpP using Abz-DFAPKMALVPYNO2 as substrate assessed as degradation of substrate peptide preincubated for 15 ... | J Nat Prod 79: 1193-7 (2016) Article DOI: 10.1021/acs.jnatprod.5b01091 BindingDB Entry DOI: 10.7270/Q2TH8R62 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| ATP-dependent Clp protease proteolytic subunit (Escherichia coli (strain K12)) | BDBM50535809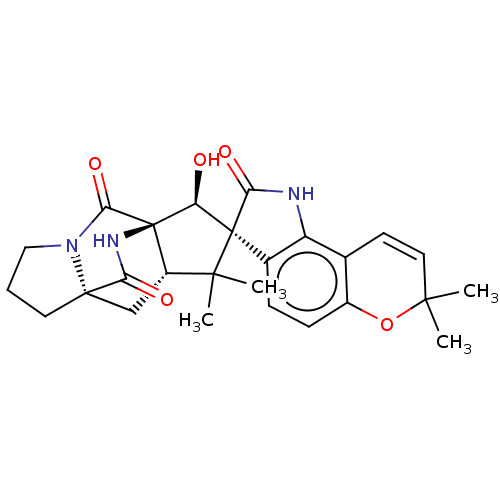 (Sclerotiamide) | PDB KEGG UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | n/a | 3.96E+4 | n/a | n/a | n/a | n/a |
University of Oklahoma COBRE in Structural Biology Curated by ChEMBL | Assay Description Activation of Escherichia coli BLR (DE3) ClpP using Abz-DFAPKMALVPYNO2 as substrate assessed as degradation of substrate peptide preincubated for 15 ... | J Nat Prod 79: 1193-7 (2016) Article DOI: 10.1021/acs.jnatprod.5b01091 BindingDB Entry DOI: 10.7270/Q2TH8R62 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| ATP-dependent Clp protease proteolytic subunit (Escherichia coli (strain K12)) | BDBM50535811 (CHEMBL4549588) | PDB KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | n/a | 1.20E+3 | n/a | n/a | n/a | n/a |
University of Oklahoma COBRE in Structural Biology Curated by ChEMBL | Assay Description Activation of Escherichia coli BLR (DE3) ClpP using FITC-beta casein as substrate assessed as degradation of substrate peptide preincubated for 15 mi... | J Nat Prod 79: 1193-7 (2016) Article DOI: 10.1021/acs.jnatprod.5b01091 BindingDB Entry DOI: 10.7270/Q2TH8R62 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| ATP-dependent Clp protease proteolytic subunit (Escherichia coli (strain K12)) | BDBM50535810 (CHEMBL4593796) | PDB KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | n/a | 2.22E+4 | n/a | n/a | n/a | n/a |
University of Oklahoma COBRE in Structural Biology Curated by ChEMBL | Assay Description Activation of Escherichia coli BLR (DE3) ClpP using FITC-beta casein as substrate assessed as degradation of substrate peptide preincubated for 15 mi... | J Nat Prod 79: 1193-7 (2016) Article DOI: 10.1021/acs.jnatprod.5b01091 BindingDB Entry DOI: 10.7270/Q2TH8R62 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| ATP-dependent Clp protease proteolytic subunit (Escherichia coli (strain K12)) | BDBM50535809 (Sclerotiamide) | PDB KEGG UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | n/a | 8.75E+4 | n/a | n/a | n/a | n/a |
University of Oklahoma COBRE in Structural Biology Curated by ChEMBL | Assay Description Activation of Escherichia coli BLR (DE3) ClpP using FITC-beta casein as substrate assessed as degradation of substrate peptide preincubated for 15 mi... | J Nat Prod 79: 1193-7 (2016) Article DOI: 10.1021/acs.jnatprod.5b01091 BindingDB Entry DOI: 10.7270/Q2TH8R62 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| ATP-dependent Clp protease proteolytic subunit (Escherichia coli (strain K12)) | BDBM50535811 (CHEMBL4549588) | PDB KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | n/a | 100 | n/a | n/a | n/a | n/a |
University of Oklahoma COBRE in Structural Biology Curated by ChEMBL | Assay Description Activation of Escherichia coli BLR (DE3) ClpP using Abz-DFAPKMALVPYNO2 as substrate assessed as degradation of substrate peptide preincubated for 15 ... | J Nat Prod 79: 1193-7 (2016) Article DOI: 10.1021/acs.jnatprod.5b01091 BindingDB Entry DOI: 10.7270/Q2TH8R62 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||