

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Cell Reactant: | FAD-Binding Domain | ||
| Syringe Reactant: | BDBM11941 | ||
| Meas. Tech.: | Isothermal Titration Calorimetry | ||
| Entry Date: | 11/07/06 | ||
| ΔG°: | -43.1± (kJ/mole) | ||
| pH: | 7±n/a | ||
| Log10Kb: | 7.45± 6.34 | ||
| Temperature: | 298.15±n/a (K) | ||
| ΔH° : | n/a | ||
| ΔHobs : | -79.4±0 (kJ/mole) | ||
| Ionic Strength: | n/a | ||
| not known | |||
| Protons Released: | n/a | ||
| ΔCp : | -1.254±n/a (kJ/mole) | ||
| Stoich. Param.: | 0.9 | ||
| ΔS° : | -0.129± (kJ/mole-K) | ||
| Comments: | n/a | ||
| Citation |  Hoffman, JM; Grunau, A; Smith, AM; Paine, MJ; Rooney, CS; Ladbury, JE; Fisher, TE; Gutierrez, A; Wai, JS; Thomas, CM; Bamberger, DL; Barnes, JL; Williams, TM; Jones, JH Global effects of the energetics of coenzyme binding: NADPH controls the protein interaction properties of human cytochrome P450 reductase. Biochemistry45:1421-34 (2006) [PubMed] Article Hoffman, JM; Grunau, A; Smith, AM; Paine, MJ; Rooney, CS; Ladbury, JE; Fisher, TE; Gutierrez, A; Wai, JS; Thomas, CM; Bamberger, DL; Barnes, JL; Williams, TM; Jones, JH Global effects of the energetics of coenzyme binding: NADPH controls the protein interaction properties of human cytochrome P450 reductase. Biochemistry45:1421-34 (2006) [PubMed] Article | ||
| More Info.: | Get all data from this article , ITC RUN data , Solution Info , Data Fit Method , Instrument Info | ||
| FAD-Binding Domain | |||
| Source: | The functional FAD-binding domain, includes the linker region, of human fibroblast CPR was expressed in Escherichia coli BL21 (DE3). | ||
| Purity: | n/a | ||
| Prep. Method: | The recombinant His-tagged proteins were purified to homogeneity by nickel-agarose chromatography. The notable exception is the omission of the 2,5-ADP affinity resin step to avoid the unusual biphasic binding isotherms during ITC experiment. | ||
| Name: | FAD-Binding Domain | ||
| Synonyms: | CYPOR | NCPR_HUMAN | POR | residue 273 to 492 of NADPH--cytochrome P450 reductase | ||
| Type: | enzyme co-factor binding domain | ||
| Mol. Mass.: | 24688.74 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P16435[273-492] | ||
| Residue: | 220 | ||
| Sequence: |
| ||
| BDBM11941 | |||
| Source: | Sigma-Aldrich | ||
| Purity: | n/a | ||
| Prep. Method: | n/a | ||
| Name | BDBM11941 | ||
| Synonyms: | 2,5-ADP | ADENOSINE-2 -5 -DIPHOSPHATE | {[(2R,3R,4R,5R)-5-(6-amino-9H-purin-9-yl)-3-hydroxy-4-(phosphonooxy)oxolan-2-yl]methoxy}phosphonic acid | ||
| Type | Nucleoside or nucleotide | ||
| Emp. Form. | C10H15N5O10P2 | ||
| Mol. Mass. | 427.2011 | ||
| SMILES | Nc1ncnc2n(cnc12)[C@@H]1O[C@H](COP(O)(O)=O)[C@@H](O)[C@H]1OP(O)(O)=O |r| | ||
| Structure | 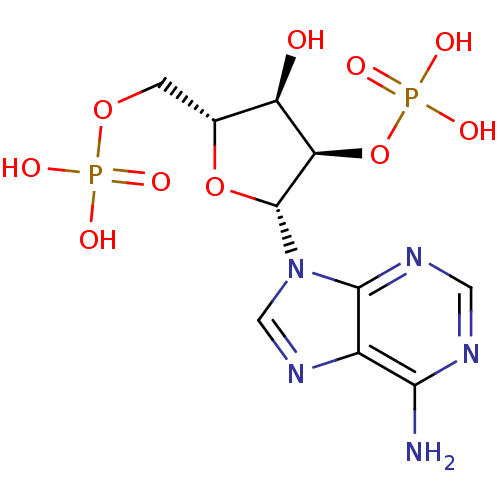
| ||
|
Home |
| |
Search |
| |
Deposit |
| |
SiteMap |
| |
About us |
| |
Email us |
| |
Info |
|
©2000 BindingDB. All rights reserved. |
|||||||||||||