

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Cell Reactant: | tRNA synthetase (GlyRS) | ||
| Syringe Reactant: | BDBM18138 | ||
| Meas. Tech.: | Isothermal Titration Calorimetry | ||
| Entry Date: | 10/01/07 | ||
| ΔG°: | -37.4±0.800 (kJ/mole) | ||
| pH: | 7.2±n/a | ||
| Log10Kb: | 6.57± 6.04 | ||
| Temperature: | 298±n/a (K) | ||
| ΔH° : | n/a | ||
| ΔHobs : | -187±9.5 (kJ/mole) | ||
| Ionic Strength: | n/a | ||
| yes | |||
| Protons Released: | n/a | ||
| ΔCp : | n/a | ||
| Stoich. Param.: | 1.05 | ||
| ΔS° : | -0.5±0.0309 (kJ/mole-K) | ||
| Comments: | n/a | ||
| Citation |  Dignam, JD; Nada, S; Chaires, JB Thermodynamic characterization of the binding of nucleotides to glycyl-tRNA synthetase. Biochemistry42:5333-40 (2003) [PubMed] Article Dignam, JD; Nada, S; Chaires, JB Thermodynamic characterization of the binding of nucleotides to glycyl-tRNA synthetase. Biochemistry42:5333-40 (2003) [PubMed] Article | ||
| More Info.: | Get all data from this article , ITC RUN data , Solution Info , Data Fit Method , Instrument Info | ||
| tRNA synthetase (GlyRS) | |||
| Source: | Protein was expressed and purified from E. coli. | ||
| Purity: | n/a | ||
| Prep. Method: | Enzyme was purified by affinity chromatography. | ||
| Name: | Glycine--tRNA ligase | ||
| Synonyms: | GARS_BOMMO | GlyRS | Glycine--tRNA ligase | Glycyl-tRNA synthetase | tRNA synthetase (GlyRS) | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 76917.70 | ||
| Organism: | Bombyx mori | ||
| Description: | B. mori glycyltRNA synthetase was expressed in E. coli. | ||
| Residue: | 680 | ||
| Sequence: |
| ||
| BDBM18138 | |||
| Source: | Sigma-Aldrich | ||
| Purity: | n/a | ||
| Prep. Method: | n/a | ||
| Name | BDBM18138 | ||
| Synonyms: | 2-amino-1-[({[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}sulfonyl)amino]ethan-1-one | 5-O-[N-(L-glycyl)sulfamoyl]adenosine | GSAd | Glycine sulfamoyl adenosine | ||
| Type | Nucleoside or nucleotide | ||
| Emp. Form. | C12H17N7O7S | ||
| Mol. Mass. | 403.371 | ||
| SMILES | NCC(=O)NS(=O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1O)n1cnc2c(N)ncnc12 |r| | ||
| Structure | 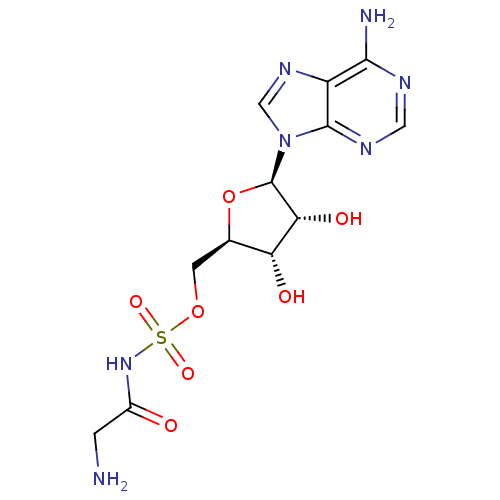
| ||
|
Home |
| |
Search |
| |
Deposit |
| |
SiteMap |
| |
About us |
| |
Email us |
| |
Info |
|
©2000 BindingDB. All rights reserved. |
|||||||||||||