

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Cell Reactant: | v-Src SH2 Domain | ||
| Syringe Reactant: | BDBM22599 | ||
| Meas. Tech.: | Isothermal Titration Calorimetry | ||
| Entry Date: | 06/14/08 | ||
| ΔG°: | -22.7± (kJ/mole) | ||
| pH: | 6±n/a | ||
| Log10Kb: | 3.98± 3.41 | ||
| Temperature: | 298.15±n/a (K) | ||
| ΔH° : | n/a | ||
| ΔHobs : | -19±0.560 (kJ/mole) | ||
| Ionic Strength: | n/a | ||
| no | |||
| Protons Released: | n/a | ||
| ΔCp : | n/a | ||
| Stoich. Param.: | n/a | ||
| ΔS° : | 0.0124± (kJ/mole-K) | ||
| Comments: | ITC data were obtained through competition experiments using high affinity peptide (PQpYEEIPI) as the competitor. | ||
| Citation |  Taylor, JD; Gilbert, PJ; Williams, MA; Pitt, WR; Ladbury, JE Identification of novel fragment compounds targeted against the pY pocket of v-Src SH2 by computational and NMR screening and thermodynamic evaluation. Proteins67:981-90 (2007) [PubMed] Article Taylor, JD; Gilbert, PJ; Williams, MA; Pitt, WR; Ladbury, JE Identification of novel fragment compounds targeted against the pY pocket of v-Src SH2 by computational and NMR screening and thermodynamic evaluation. Proteins67:981-90 (2007) [PubMed] Article | ||
| More Info.: | Get all data from this article , ITC RUN data , Solution Info , Data Fit Method , Instrument Info | ||
| v-Src SH2 Domain | |||
| Source: | n/a | ||
| Purity: | n/a | ||
| Prep. Method: | n/a | ||
| Name: | Tyrosine-protein kinase transforming protein Src | ||
| Synonyms: | SRC_RSVSA | V-SRC | p60-Src | v-Src SH2 Domain | ||
| Type: | SH2 Domain | ||
| Mol. Mass.: | 58884.32 | ||
| Organism: | Avian leukosis virus (RSA) | ||
| Description: | P00524 | ||
| Residue: | 526 | ||
| Sequence: |
| ||
| BDBM22599 | |||
| Source: | n/a | ||
| Purity: | n/a | ||
| Prep. Method: | n/a | ||
| Name | BDBM22599 | ||
| Synonyms: | 5-bromo-4-chloro-3-indoxyl phosphate | Fragment compound, 6 | [(5-bromo-4-chloro-1H-indol-3-yl)oxy]phosphonic acid | ||
| Type | Small organic molecule | ||
| Emp. Form. | C8H6BrClNO4P | ||
| Mol. Mass. | 326.468 | ||
| SMILES | OP(O)(=O)Oc1c[nH]c2ccc(Br)c(Cl)c12 | ||
| Structure | 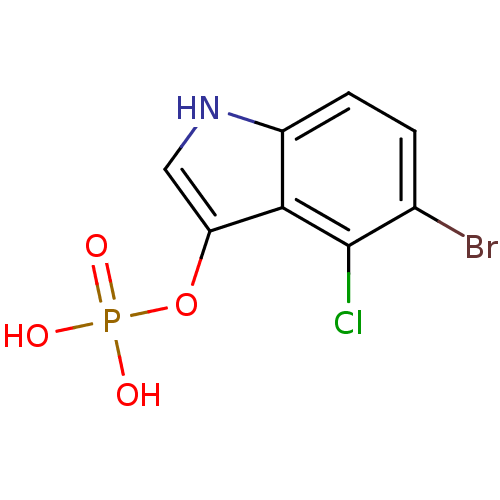
| ||
|
Home |
| |
Search |
| |
Deposit |
| |
SiteMap |
| |
About us |
| |
Email us |
| |
Info |
|
©2000 BindingDB. All rights reserved. |
|||||||||||||