

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Cell Reactant: | Serine Hydroxymethyltransferase | ||
| Syringe Reactant: | BDBM289 | ||
| Meas. Tech.: | Isothermal Titration Calorimetry | ||
| Entry Date: | 03/31/03 | ||
| ΔG°: | -10.1± (kcal/mole) | ||
| pH: | n/a | ||
| Log10Kb: | 7.46± n/a | ||
| Temperature: | 298.15±n/a (K) | ||
| ΔH° : | 7± (kcal/mole) | ||
| ΔHobs : | 7± (kcal/mole) | ||
| Ionic Strength: | n/a | ||
| not known | |||
| Protons Released: | n/a | ||
| ΔCp : | -0.43±n/a (kcal/mole) | ||
| Stoich. Param.: | n/a | ||
| ΔS° : | n/a | ||
| Comments: | n/a | ||
| Citation |  Hom, RK; Huang, T; Gailunas, AF; Wang, C; Mamo, S; Maras, B; Fang, LY; Barra, D; Tung, JS; Walker, DE; Schirch, V; Davis, D; Thorsett, ED; Jewett, NE; Moon, JB; John, V Thermodynamic analysis of the binding of the polyglutamate chain of 5-formyltetrahydropteroylpolyglutamates to serine hydroxymethyltransferase. Biochemistry37:13536-42 (1998) [PubMed] Article Hom, RK; Huang, T; Gailunas, AF; Wang, C; Mamo, S; Maras, B; Fang, LY; Barra, D; Tung, JS; Walker, DE; Schirch, V; Davis, D; Thorsett, ED; Jewett, NE; Moon, JB; John, V Thermodynamic analysis of the binding of the polyglutamate chain of 5-formyltetrahydropteroylpolyglutamates to serine hydroxymethyltransferase. Biochemistry37:13536-42 (1998) [PubMed] Article | ||
| More Info.: | Get all data from this article , ITC RUN data , Solution Info , Data Fit Method , Instrument Info | ||
| Serine Hydroxymethyltransferase | |||
| Source: | purified in the laboratory | ||
| Purity: | n/a | ||
| Prep. Method: | SHMT, about 6 mg/mL, was dialyzed overnight against 20 mM potassium phosphate buffer at pH 7.0, containing 50 mM glycine and 1 mM DTT. This enzyme was then diluted to the desired concentration with the dialysis buffer. | ||
| Name: | Serine Hydroxymethyltransferase | ||
| Synonyms: | GLYC_RABIT | SHMT1 | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 52984.86 | ||
| Organism: | Oryctolagus cuniculus (rabbit) | ||
| Description: | P07511 | ||
| Residue: | 484 | ||
| Sequence: |
| ||
| BDBM289 | |||
| Source: | B. Schircks | ||
| Purity: | n/a | ||
| Prep. Method: | A concentrated solution of 5-CHO-H4PteGlun was passed through a 1 cm x 10 cm Bio-Gel P-2 column equilibrated with dialysis buffer. The concentration of the solution was determined from its absorbance at 288 nm using an extinction coefficient of 31 500 M -1 cm -1 | ||
| Name | BDBM289 | ||
| Synonyms: | (2S)-2-[(2S)-2-[(2S)-2-[(2S)-2-{[4-({[(6S)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]formamido}-4-formamidobutanoic acid]-4-formamidobutanoic acid]-4-formamidobutanoic acid]pentanedioic acid | 5-CHO-H4PteGlu4 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C35H44N10O16 | ||
| Mol. Mass. | 860.7813 | ||
| SMILES | Nc1nc2NC[C@H](CNc3ccc(cc3)C(=O)N[C@@H](CCC(=O)N[C@@H](CCC(=O)N[C@@H](CCC(=O)N[C@@H](CCC(O)=O)C(O)=O)C(O)=O)C(O)=O)C(O)=O)N(C=O)c2c(=O)[nH]1 | ||
| Structure | 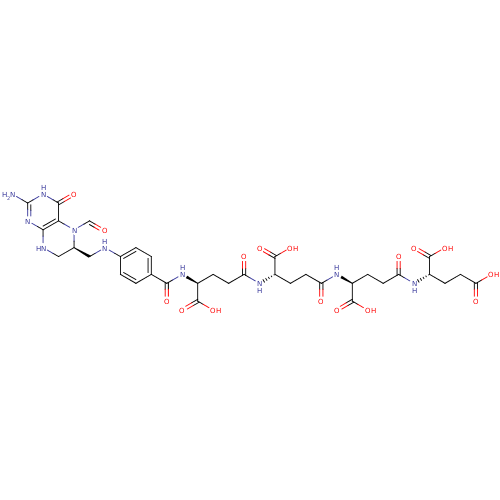
| ||
|
Home |
| |
Search |
| |
Deposit |
| |
SiteMap |
| |
About us |
| |
Email us |
| |
Info |
|
©2000 BindingDB. All rights reserved. |
|||||||||||||