

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Cell Reactant: | HIV-1 Protease B Subtype | ||
| Syringe Reactant: | BDBM580 | ||
| Meas. Tech.: | Isothermal Titration Calorimetry | ||
| Entry Date: | 06/22/04 | ||
| ΔG°: | -62.3± (kJ/mole) | ||
| pH: | 5±n/a | ||
| Log10Kb: | 11.0± 9.62 | ||
| Temperature: | 298.15±n/a (K) | ||
| ΔH° : | -33.4± (kJ/mole) | ||
| ΔHobs : | -33.4± (kJ/mole) | ||
| Ionic Strength: | n/a | ||
| not known | |||
| Protons Released: | n/a | ||
| ΔCp : | n/a | ||
| Stoich. Param.: | n/a | ||
| ΔS° : | 0.0968± (kJ/mole-K) | ||
| Comments: | In displacement titration, inhibitors were injected into the calorimetric cell containing the protein prebound to a weak inhibitor acetylpepstatin. | ||
| Citation |  Velazquez-Campoy, A; Vega, S; Freire, E Amplification of the effects of drug resistance mutations by background polymorphisms in HIV-1 protease from African subtypes. Biochemistry41:8613-9 (2002) [PubMed] Article Velazquez-Campoy, A; Vega, S; Freire, E Amplification of the effects of drug resistance mutations by background polymorphisms in HIV-1 protease from African subtypes. Biochemistry41:8613-9 (2002) [PubMed] Article | ||
| More Info.: | Get all data from this article , ITC RUN data , Solution Info , Instrument Info | ||
| HIV-1 Protease B Subtype | |||
| Source: | Plasmid-encoded HIV-1 protease was expressed in E. coli cells. | ||
| Purity: | 99% | ||
| Prep. Method: | HIV-1 protease was purified and refolded from E. coli inclusion bodies. | ||
| Name: | HIV-1 Protease B Subtype | ||
| Synonyms: | n/a | ||
| Type: | Protein Complex | ||
| Mol. Mass.: | n/a | ||
| Description: | n/a | ||
| Components: | This complex has 2 components. | ||
| Component 1 | |||
| Name: | Gag-Pol polyprotein [489-587,Q496K] | ||
| Synonyms: | HIV-1 Protease (Q7K) chain A | HIV-1 Protease (Q7K) chain B | HIV-1 Protease B Subtype chain A | HIV-1 Protease B Subtype chain B | HIV-1 Protease Mutant (Q7K) | HIV-1 Protease chain A | POL_HV1H2 | gag-pol | ||
| Type: | Enzyme Subunit | ||
| Mol. Mass.: | 10782.21 | ||
| Organism: | Human immunodeficiency virus type 1 | ||
| Description: | All of the protease constructs in this study are on the wild type containing the single Q7K mutation to prevent autocatalysis.This protein has enzymatic properties similar to that of the wild type protease. | ||
| Residue: | 99 | ||
| Sequence: |
| ||
| Component 2 | |||
| Name: | Gag-Pol polyprotein [489-587,Q496K] | ||
| Synonyms: | HIV-1 Protease (Q7K) chain A | HIV-1 Protease (Q7K) chain B | HIV-1 Protease B Subtype chain A | HIV-1 Protease B Subtype chain B | HIV-1 Protease Mutant (Q7K) | HIV-1 Protease chain A | POL_HV1H2 | gag-pol | ||
| Type: | Enzyme Subunit | ||
| Mol. Mass.: | 10782.21 | ||
| Organism: | Human immunodeficiency virus type 1 | ||
| Description: | All of the protease constructs in this study are on the wild type containing the single Q7K mutation to prevent autocatalysis.This protein has enzymatic properties similar to that of the wild type protease. | ||
| Residue: | 99 | ||
| Sequence: |
| ||
| BDBM580 | |||
| Source: | From Dr. Kiso, Department of Medicinal Chemistry, Kyoto Pharmaceutical University, Kyoto, Japan). | ||
| Purity: | n/a | ||
| Prep. Method: | were used without further purification | ||
| Name | BDBM580 | ||
| Synonyms: | (4R)-3-[(2S,3S)-2-hydroxy-3-[(3-hydroxy-2-methylphenyl)formamido]-4-phenylbutanoyl]-5,5-dimethyl-N-[(2-methylphenyl)methyl]-1,3-thiazolidine-4-carboxamide | AG1776 | BDBM50288366 | JE-2147 | KNI-764 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C32H37N3O5S | ||
| Mol. Mass. | 575.718 | ||
| SMILES | Cc1ccccc1CNC(=O)[C@H]1N(CSC1(C)C)C(=O)[C@@H](O)[C@H](Cc1ccccc1)NC(=O)c1cccc(O)c1C |r| | ||
| Structure | 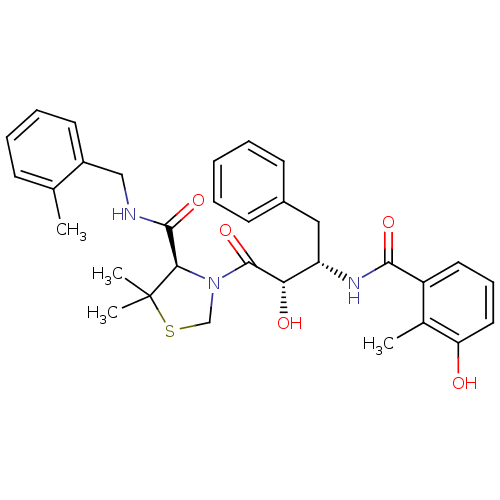
| ||
|
Home |
| |
Search |
| |
Deposit |
| |
SiteMap |
| |
About us |
| |
Email us |
| |
Info |
|
©2000 BindingDB. All rights reserved. |
|||||||||||||