

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Cell Reactant: | Enoyl-ACP Reductase (FabI) | ||
| Syringe Reactant: | BDBM3910 | ||
| Meas. Tech.: | Isothermal Titration Calorimetry | ||
| Entry Date: | 04/13/05 | ||
| ΔG°: | -41.7± (kJ/mole) | ||
| pH: | 7.5±n/a | ||
| Log10Kb: | 7.55± 6.43 | ||
| Temperature: | 288.15±n/a (K) | ||
| ΔH° : | -42.5±0.800 (kJ/mole) | ||
| ΔHobs : | -42.5±0.800 (kJ/mole) | ||
| Ionic Strength: | n/a | ||
| not known | |||
| Protons Released: | n/a | ||
| ΔCp : | -3.69±0.67 (kJ/mole) | ||
| Stoich. Param.: | 0.54 | ||
| ΔS° : | -0.0026± (kJ/mole-K) | ||
| Comments: | n/a | ||
| Citation |  Protasevich, II; Brouillette, CG; Snow, ME; Dunham, S; Rubin, JR; Gogliotti, R; Siegel, K Role of inhibitor aliphatic chain in the thermodynamics of inhibitor binding to Escherichia coli enoyl-ACP reductase and the Phe203Leu mutant: a proposed mechanism for drug resistance. Biochemistry43:13380-9 (2004) [PubMed] Article Protasevich, II; Brouillette, CG; Snow, ME; Dunham, S; Rubin, JR; Gogliotti, R; Siegel, K Role of inhibitor aliphatic chain in the thermodynamics of inhibitor binding to Escherichia coli enoyl-ACP reductase and the Phe203Leu mutant: a proposed mechanism for drug resistance. Biochemistry43:13380-9 (2004) [PubMed] Article | ||
| More Info.: | Get all data from this article , ITC RUN data , Solution Info , Data Fit Method , Instrument Info | ||
| Enoyl-ACP Reductase (FabI) | |||
| Source: | The His-tagged FabI was expressed and purified from E. coli | ||
| Purity: | Purity was not lower than 95% | ||
| Prep. Method: | The calorimetric titration of FabI was conducted in the presence of 0.5 mM NAD+. | ||
| Name: | Enoyl-ACP Reductase (FabI) | ||
| Synonyms: | n/a | ||
| Type: | Binary complex with a cofactor | ||
| Mol. Mass.: | n/a | ||
| Description: | n/a | ||
| Components: | This complex has 2 components. | ||
| Component 1 | |||
| Name: | Enoyl-ACP Reductase (FabI) | ||
| Synonyms: | NADPH-dependent enoyl-ACP reductase | ||
| Type: | Enzyme Subunit | ||
| Mol. Mass.: | 27729.92 | ||
| Organism: | Escherichia coli | ||
| Description: | P0AEK4[2-262] | ||
| Residue: | 261 | ||
| Sequence: |
| ||
| Component 2 | |||
| Name | BDBM3909 | ||
| Synonyms: | NAD+ | ||
| Type | Nucleoside or nucleotide | ||
| Emp. Form. | C21H27N7O14P2 | ||
| Mol. Mass. | 663.4251 | ||
| SMILES | NC(=O)c1ccc[n+](c1)[C@@H]1O[C@H](COP([O-])(=O)OP(O)(=O)OC[C@H]2O[C@H]([C@H](O)[C@@H]2O)n2cnc3c(N)ncnc23)[C@@H](O)[C@H]1O |r| | ||
| Structure | 
| ||
| BDBM3910 | |||
| Source: | Organic Synthesis | ||
| Purity: | n/a | ||
| Prep. Method: | n/a | ||
| Name | BDBM3910 | ||
| Synonyms: | 2-(2-hydroxyphenoxy)phenol | PD048890 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C12H10O3 | ||
| Mol. Mass. | 202.206 | ||
| SMILES | Oc1ccccc1Oc1ccccc1O | ||
| Structure | 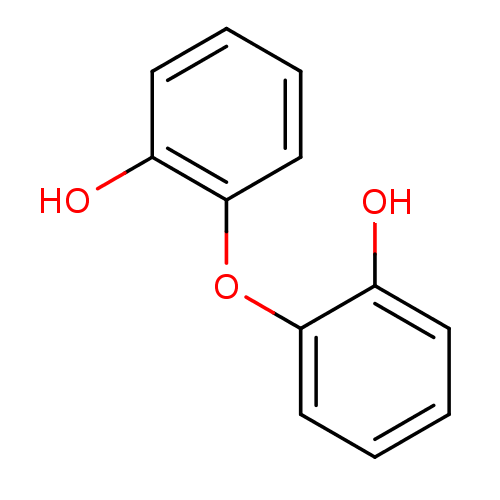
| ||
|
Home |
| |
Search |
| |
Deposit |
| |
SiteMap |
| |
About us |
| |
Email us |
| |
Info |
|
©2000 BindingDB. All rights reserved. |
|||||||||||||