

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Cell Reactant: | Haspin (465-798) | ||
| Syringe Reactant: | BDBM103302 | ||
| Meas. Tech.: | Enzyme Inhibition | ||
| Entry Date: | 10/22/14 | ||
| ΔG°: | -8.67± (kcal/mole) | ||
| pH: | 7.5±0 | ||
| Log10Kb: | 6.57± n/a | ||
| Temperature: | 288.15±0 (K) | ||
| ΔH° : | -17.6±0.100 (kcal/mole) | ||
| ΔHobs: | n/a | ||
| Ionic Strength: | n/a | ||
| n/a | |||
| Protons Released: | n/a | ||
| ΔCp : | n/a | ||
| Stoich. Param.: | n/a | ||
| ΔS° : | -0.0300±0 (kcal/mole-K) | ||
| Comments: | n/a | ||
| Citation |  Chaikuad, A; Tacconi, EM; Zimmer, J; Liang, Y; Gray, NS; Tarsounas, M; Knapp, S A unique inhibitor binding site in ER K1/2 is associated with slow binding kinetics Nat Chem Biol10:853-860 (2014) [PubMed] Article Chaikuad, A; Tacconi, EM; Zimmer, J; Liang, Y; Gray, NS; Tarsounas, M; Knapp, S A unique inhibitor binding site in ER K1/2 is associated with slow binding kinetics Nat Chem Biol10:853-860 (2014) [PubMed] Article | ||
| More Info.: | Get all data from this article , ITC RUN data | ||
| Haspin (465-798) | |||
| Source: | n/a | ||
| Purity: | n/a | ||
| Prep. Method: | n/a | ||
| Name: | Serine/threonine-protein kinase haspin [465-798] | ||
| Synonyms: | GSG2 | HASPIN | HASP_HUMAN | Haspin (aa 465-798) | ||
| Type: | Protein | ||
| Mol. Mass.: | 37973.67 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Haspin kinase domain 465-798aa (curated by PDB 4QTC) | ||
| Residue: | 334 | ||
| Sequence: |
| ||
| BDBM103302 | |||
| Source: | n/a | ||
| Purity: | n/a | ||
| Prep. Method: | n/a | ||
| Name | BDBM103302 | ||
| Synonyms: | SCH772984 | US8546404, 6 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C33H33N9O2 | ||
| Mol. Mass. | 587.6742 | ||
| SMILES | O=C(CN1CC[C@@H](C1)C(=O)Nc1ccc2[nH]nc(-c3ccncc3)c2c1)N1CCN(CC1)c1ccc(cc1)-c1ncccn1 |r| | ||
| Structure | 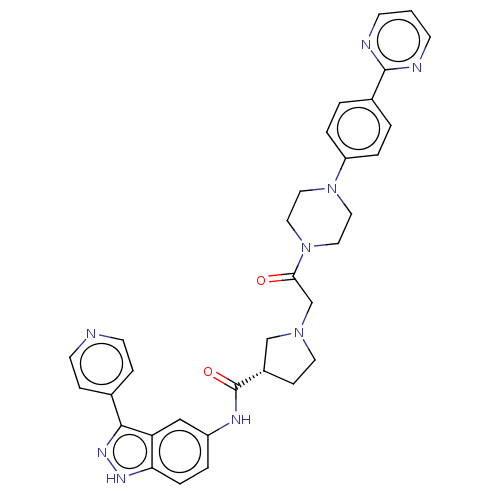
| ||
|
Home |
| |
Search |
| |
Deposit |
| |
SiteMap |
| |
About us |
| |
Email us |
| |
Info |
|
©2000 BindingDB. All rights reserved. |
|||||||||||||