

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Cell Reactant: | Methylthioadenosine Phosphorylase (MTAP) | ||
| Syringe Reactant: | BDBM218798 | ||
| Meas. Tech.: | Enzyme Inhibition | ||
| Entry Date: | 03/22/17 | ||
| Δ G ° : | n/a | ||
| pH: | 7.4±0 | ||
| Log10Kb: | n/a | ||
| Temperature: | 310.15±0 (K) | ||
| ΔH° : | -16±0.400 (kJ/mole) | ||
| ΔHobs: | n/a | ||
| Ionic Strength: | n/a | ||
| n/a | |||
| Protons Released: | n/a | ||
| ΔCp : | n/a | ||
| Stoich. Param.: | n/a | ||
| ΔS° : | n/a | ||
| Comments: | n/a | ||
| Citation |  Firestone, RS; Cameron, SA; Karp, JM; Arcus, VL; Schramm, VL Heat Capacity Changes for Transition-State Analogue Binding and Catalysis with Human 5'-Methylthioadenosine Phosphorylase. ACS Chem Biol12:464-473 (2017) [PubMed] Article Firestone, RS; Cameron, SA; Karp, JM; Arcus, VL; Schramm, VL Heat Capacity Changes for Transition-State Analogue Binding and Catalysis with Human 5'-Methylthioadenosine Phosphorylase. ACS Chem Biol12:464-473 (2017) [PubMed] Article | ||
| More Info.: | Get all data from this article , ITC RUN data , Instrument Info | ||
| Methylthioadenosine Phosphorylase (MTAP) | |||
| Source: | n/a | ||
| Purity: | n/a | ||
| Prep. Method: | n/a | ||
| Name: | S-methyl-5'-thioadenosine phosphorylase | ||
| Synonyms: | 5'-Methylthioadenosine phosphorylase (MTAP) | MSAP | MTA phosphorylase | MTAP | MTAP_HUMAN | MTAPase | Methylthioadenosine Phosphorylase (MTAP) | S-methyl-5 -thioadenosine phosphorylase | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 31239.23 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | n/a | ||
| Residue: | 283 | ||
| Sequence: |
| ||
| BDBM218798 | |||
| Source: | n/a | ||
| Purity: | n/a | ||
| Prep. Method: | n/a | ||
| Name | BDBM218798 | ||
| Synonyms: | MT-DADMe-ImmA | ||
| Type | Small organic molecule | ||
| Emp. Form. | C13H20N5OS | ||
| Mol. Mass. | 294.395 | ||
| SMILES | CSC[C@H]1C[NH+](Cc2c[nH]c3c(N)ncnc23)C[C@@H]1O | ||
| Structure | 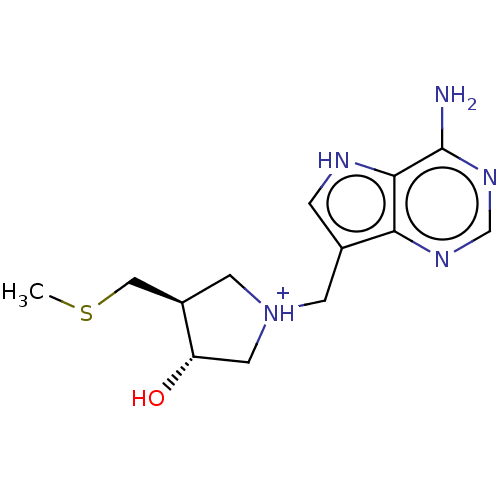
| ||
|
Home |
| |
Search |
| |
Deposit |
| |
SiteMap |
| |
About us |
| |
Email us |
| |
Info |
|
©2000 BindingDB. All rights reserved. |
|||||||||||||