

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Cell Reactant: | Phospholipase C-γ1 SH2 domain (PLCC_R716E) | ||
| Syringe Reactant: | BDBM225241 | ||
| Meas. Tech.: | Enzyme Inhibition | ||
| Entry Date: | 05/11/17 | ||
| ΔG°: | -10.7± (kcal/mole) | ||
| pH: | 7.4±0 | ||
| Log10Kb: | 7.86± 7.22 | ||
| Temperature: | 298.15±0 (K) | ||
| ΔH° : | n/a | ||
| ΔHobs: | n/a | ||
| Ionic Strength: | n/a | ||
| n/a | |||
| Protons Released: | n/a | ||
| ΔCp : | n/a | ||
| Stoich. Param.: | n/a | ||
| ΔS° : | n/a | ||
| Comments: | n/a | ||
| Citation |  McKercher, MA; Guan, X; Tan, Z; Wuttke, DS Multimodal Recognition of Diverse Peptides by the C-Terminal SH2 Domain of Phospholipase C-¿1 Protein. Biochemistry56:2225-2237 (2017) [PubMed] Article McKercher, MA; Guan, X; Tan, Z; Wuttke, DS Multimodal Recognition of Diverse Peptides by the C-Terminal SH2 Domain of Phospholipase C-¿1 Protein. Biochemistry56:2225-2237 (2017) [PubMed] Article | ||
| More Info.: | Get all data from this article , ITC RUN data | ||
| Phospholipase C-γ1 SH2 domain (PLCC_R716E) | |||
| Source: | n/a | ||
| Purity: | n/a | ||
| Prep. Method: | n/a | ||
| Name: | Phospholipase C-γ1 SH2 domain (PLCC_R716E) | ||
| Synonyms: | n/a | ||
| Type: | Protein | ||
| Mol. Mass.: | 11531.50 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Human phospholipase C-γ1 C-terminal SH2 domain (663-759 aa) R716E mutant | ||
| Residue: | 97 | ||
| Sequence: |
| ||
| BDBM225241 | |||
| Source: | n/a | ||
| Purity: | n/a | ||
| Prep. Method: | n/a | ||
| Name | BDBM225241 | ||
| Synonyms: | 3-Nitro-N-(2-phenylbenzo[d]oxazol-6-yl)benzenesulfonamide (2) | MT-pep | ||
| Type | Small organic molecule | ||
| Emp. Form. | C63H91N12O24P | ||
| Mol. Mass. | 1431.4364 | ||
| SMILES | CC[C@H](C)[C@H](NC(=O)[C@@H]1CCCN1C(=O)[C@@H](NC(=O)[C@H](CCC(O)=O)NC(=O)[C@H](CCC(O)=O)NC(=O)[C@H](Cc1ccc(OP(O)(O)=O)cc1)NC(=O)[C@H](CCC(N)=O)NC(=O)[C@@H]1CCCN1C(=O)[C@H](CCC(O)=O)NC(=O)[C@H](C)N)[C@@H](C)CC)C(=O)N[C@@H](Cc1ccc(O)cc1)C(O)=O |r| | ||
| Structure | 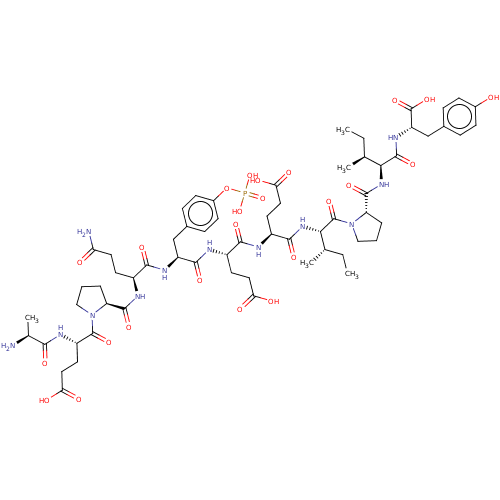
| ||
|
Home |
| |
Search |
| |
Deposit |
| |
SiteMap |
| |
About us |
| |
Email us |
| |
Info |
|
©2000 BindingDB. All rights reserved. |
|||||||||||||