

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Cell Reactant: | Nuclear receptor REV-ERBβ ligand-binding domain (REV-ERBβ-LBD) | ||
| Syringe Reactant: | BDBM231645 | ||
| Meas. Tech.: | Enzyme Inhibition | ||
| Entry Date: | 06/06/17 | ||
| ΔG°: | -36.8± (kJ/mole) | ||
| pH: | 7.5±0 | ||
| Log10Kb: | 6.45± 6.45 | ||
| Temperature: | 298.15±0 (K) | ||
| ΔH° : | -104±0 (kJ/mole) | ||
| ΔHobs: | n/a | ||
| Ionic Strength: | n/a | ||
| n/a | |||
| Protons Released: | n/a | ||
| ΔCp : | n/a | ||
| Stoich. Param.: | n/a | ||
| ΔS° : | -0.232±0 (kJ/mole-K) | ||
| Comments: | n/a | ||
| Citation |  Matta-Camacho, E; Banerjee, S; Hughes, TS; Solt, LA; Wang, Y; Burris, TP; Kojetin, DJ Structure of REV-ERB▀ ligand-binding domain bound to a porphyrin antagonist. J Biol Chem289:20054-66 (2014) [PubMed] Article Matta-Camacho, E; Banerjee, S; Hughes, TS; Solt, LA; Wang, Y; Burris, TP; Kojetin, DJ Structure of REV-ERB▀ ligand-binding domain bound to a porphyrin antagonist. J Biol Chem289:20054-66 (2014) [PubMed] Article | ||
| More Info.: | Get all data from this article , ITC RUN data | ||
| Nuclear receptor REV-ERBβ ligand-binding domain (REV-ERBβ-LBD) | |||
| Source: | n/a | ||
| Purity: | n/a | ||
| Prep. Method: | n/a | ||
| Name: | Nuclear receptor REV-ERB╬▓ ligand-binding domain (REV-ERB╬▓-LBD) | ||
| Synonyms: | NR1D2 | NR1D2_HUMAN | ||
| Type: | Protein | ||
| Mol. Mass.: | 22445.55 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Human REV-ERB╬▓-LBD (381-579 aa) | ||
| Residue: | 199 | ||
| Sequence: |
| ||
| BDBM231645 | |||
| Source: | n/a | ||
| Purity: | n/a | ||
| Prep. Method: | n/a | ||
| Name | BDBM231645 | ||
| Synonyms: | Heme | ||
| Type | Small organic molecule | ||
| Emp. Form. | C34H32FeN4O4 | ||
| Mol. Mass. | 616.487 | ||
| SMILES | CC1=C(CCC(O)=O)C2=[N]3C1=Cc1c(C=C)c(C)c4C=C5C(C=C)=C(C)C6=[N]5[Fe]3(n14)n1c(=C6)c(C)c(CCC(O)=O)c1=C2 |c:1,8,27,36,48,t:11,20,24| | ||
| Structure | 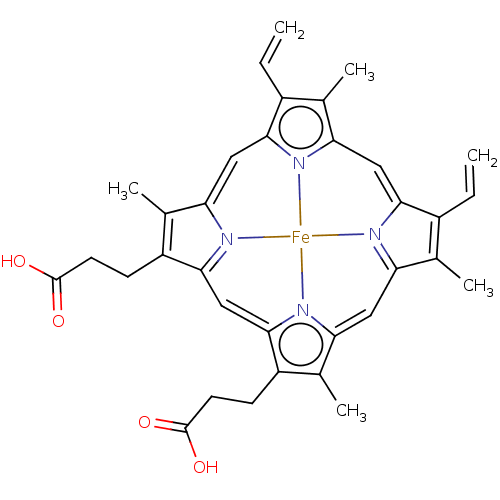
| ||
|
Home |
| |
Search |
| |
Deposit |
| |
SiteMap |
| |
About us |
| |
Email us |
| |
Info |
|
©2000 BindingDB. All rights reserved. |
|||||||||||||