| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Serine protease 1 |
|---|
| Ligand | BDBM12596 |
|---|
| Substrate/Competitor | BDBM12500 |
|---|
| Meas. Tech. | Enzyme Assay and Determination of the Inhibition Constants. |
|---|
| pH | 7.5±n/a |
|---|
| Temperature | 295.15±n/a K |
|---|
| Ki | >2900±n/a nM |
|---|
| Citation |  Maignan, S; Guilloteau, JP; Choi-Sledeski, YM; Becker, MR; Ewing, WR; Pauls, HW; Spada, AP; Mikol, V Molecular structures of human factor Xa complexed with ketopiperazine inhibitors: preference for a neutral group in the S1 pocket. J Med Chem46:685-90 (2003) [PubMed] Article Maignan, S; Guilloteau, JP; Choi-Sledeski, YM; Becker, MR; Ewing, WR; Pauls, HW; Spada, AP; Mikol, V Molecular structures of human factor Xa complexed with ketopiperazine inhibitors: preference for a neutral group in the S1 pocket. J Med Chem46:685-90 (2003) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Inhibition_Run data, Solution Info, Assay Method |
|---|
| |
| Serine protease 1 |
|---|
| Name: | Serine protease 1 |
|---|
| Synonyms: | Beta-Trypsin | Cationic trypsin | PRSS1 | TRP1 | TRY1 | TRY1_BOVIN | TRYP1 | Trypsin | Trypsin I |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 25790.52 |
|---|
| Organism: | Bos taurus (bovine) |
|---|
| Description: | P00760 |
|---|
| Residue: | 246 |
|---|
| Sequence: | MKTFIFLALLGAAVAFPVDDDDKIVGGYTCGANTVPYQVSLNSGYHFCGGSLINSQWVVS
AAHCYKSGIQVRLGEDNINVVEGNEQFISASKSIVHPSYNSNTLNNDIMLIKLKSAASLN
SRVASISLPTSCASAGTQCLISGWGNTKSSGTSYPDVLKCLKAPILSDSSCKSAYPGQIT
SNMFCAGYLEGGKDSCQGDSGGPVVCSGKLQGIVSWGSGCAQKNKPGVYTKVCNYVSWIK
QTIASN
|
|
|
|---|
| BDBM12596 |
|---|
| BDBM12500 |
|---|
| Name | BDBM12596 |
|---|
| Synonyms: | 4-{[(E)-2-(5-CHLOROTHIEN-2-YL)VINYL]SULFONYL}-1-(1H-PYRROLO[3,2-C]PYRIDIN-2-YLMETHYL)PIPERAZIN-2-ONE | 4-{[(E)-2-(5-chlorothiophen-2-yl)ethene]sulfonyl}-1-{1H-pyrrolo[3,2-c]pyridin-2-ylmethyl}piperazin-2-one | CHEMBL423417 | Ketopiperazine | RPR209685 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C18H17ClN4O3S2 |
|---|
| Mol. Mass. | 436.936 |
|---|
| SMILES | Clc1ccc(C=CS(=O)(=O)N2CCN(Cc3cc4cnccc4[nH]3)C(=O)C2)s1 |w:5.4| |
|---|
| Structure | 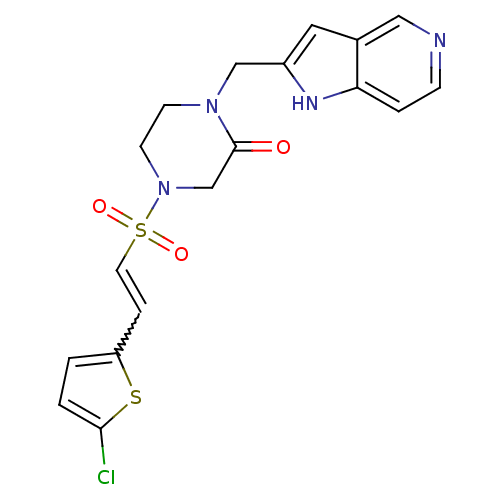
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Maignan, S; Guilloteau, JP; Choi-Sledeski, YM; Becker, MR; Ewing, WR; Pauls, HW; Spada, AP; Mikol, V Molecular structures of human factor Xa complexed with ketopiperazine inhibitors: preference for a neutral group in the S1 pocket. J Med Chem46:685-90 (2003) [PubMed] Article
Maignan, S; Guilloteau, JP; Choi-Sledeski, YM; Becker, MR; Ewing, WR; Pauls, HW; Spada, AP; Mikol, V Molecular structures of human factor Xa complexed with ketopiperazine inhibitors: preference for a neutral group in the S1 pocket. J Med Chem46:685-90 (2003) [PubMed] Article