| Reaction Details |
|---|
 | Report a problem with these data |
| Target | AP2-associated protein kinase 1 |
|---|
| Ligand | BDBM373556 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Kinase Assay |
|---|
| IC50 | 0.670±n/a nM |
|---|
| Citation |  Luo, G; Chen, L; Dzierba, CD; Ditta, JL; Macor, JE; Bronson, JJ Biaryl kinase inhibitors US Patent US9902722 Publication Date 2/27/2018 Luo, G; Chen, L; Dzierba, CD; Ditta, JL; Macor, JE; Bronson, JJ Biaryl kinase inhibitors US Patent US9902722 Publication Date 2/27/2018 |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| AP2-associated protein kinase 1 |
|---|
| Name: | AP2-associated protein kinase 1 |
|---|
| Synonyms: | AAK1 | AAK1_HUMAN | Adaptor-associated kinase 1 | KIAA1048 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 103884.23 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_774569 |
|---|
| Residue: | 961 |
|---|
| Sequence: | MKKFFDSRREQGGSGLGSGSSGGGGSTSGLGSGYIGRVFGIGRQQVTVDEVLAEGGFAIV
FLVRTSNGMKCALKRMFVNNEHDLQVCKREIQIMRDLSGHKNIVGYIDSSINNVSSGDVW
EVLILMDFCRGGQVVNLMNQRLQTGFTENEVLQIFCDTCEAVARLHQCKTPIIHRDLKVE
NILLHDRGHYVLCDFGSATNKFQNPQTEGVNAVEDEIKKYTTLSYRAPEMVNLYSGKIIT
TKADIWALGCLLYKLCYFTLPFGESQVAICDGNFTIPDNSRYSQDMHCLIRYMLEPDPDK
RPDIYQVSYFSFKLLKKECPIPNVQNSPIPAKLPEPVKASEAAAKKTQPKARLTDPIPTT
ETSIAPRQRPKAGQTQPNPGILPIQPALTPRKRATVQPPPQAAGSSNQPGLLASVPQPKP
QAPPSQPLPQTQAKQPQAPPTPQQTPSTQAQGLPAQAQATPQHQQQLFLKQQQQQQQPPP
AQQQPAGTFYQQQQAQTQQFQAVHPATQKPAIAQFPVVSQGGSQQQLMQNFYQQQQQQQQ
QQQQQQLATALHQQQLMTQQAALQQKPTMAAGQQPQPQPAAAPQPAPAQEPAIQAPVRQQ
PKVQTTPPPAVQGQKVGSLTPPSSPKTQRAGHRRILSDVTHSAVFGVPASKSTQLLQAAA
AEASLNKSKSATTTPSGSPRTSQQNVYNPSEGSTWNPFDDDNFSKLTAEELLNKDFAKLG
EGKHPEKLGGSAESLIPGFQSTQGDAFATTSFSAGTAEKRKGGQTVDSGLPLLSVSDPFI
PLQVPDAPEKLIEGLKSPDTSLLLPDLLPMTDPFGSTSDAVIEKADVAVESLIPGLEPPV
PQRLPSQTESVTSNRTDSLTGEDSLLDCSLLSNPTTDLLEEFAPTAISAPVHKAAEDSNL
ISGFDVPEGSDKVAEDEFDPIPVLITKNPQGGHSRNSSGSSESSLPNLARSLLLVDQLID
L
|
|
|
|---|
| BDBM373556 |
|---|
| n/a |
|---|
| Name | BDBM373556 |
|---|
| Synonyms: | US9902722, Example 29 | methyl (4-(4-((2-amino-2,4-dimethylpentyl)oxy)-3-chlorophenyl)pyridin-2-yl)carbamate |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H26ClN3O3 |
|---|
| Mol. Mass. | 391.892 |
|---|
| SMILES | COC(=O)Nc1cc(ccn1)-c1ccc(OCC(C)(N)CC(C)C)c(Cl)c1 |
|---|
| Structure | 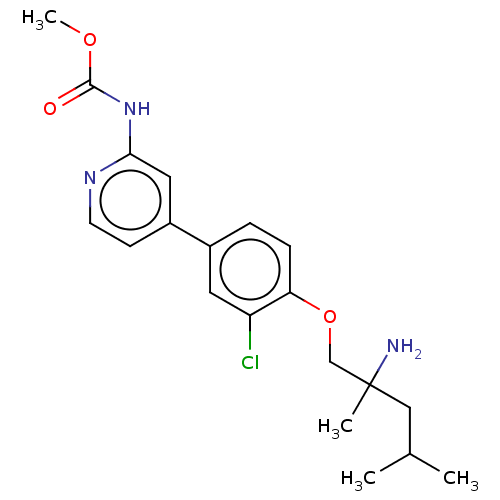
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Luo, G; Chen, L; Dzierba, CD; Ditta, JL; Macor, JE; Bronson, JJ Biaryl kinase inhibitors US Patent US9902722 Publication Date 2/27/2018
Luo, G; Chen, L; Dzierba, CD; Ditta, JL; Macor, JE; Bronson, JJ Biaryl kinase inhibitors US Patent US9902722 Publication Date 2/27/2018