| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Epoxide hydrolase B |
|---|
| Ligand | BDBM25728 |
|---|
| Substrate/Competitor | BDBM25726 |
|---|
| Meas. Tech. | Mtb EHB Inhibition Assay |
|---|
| pH | 7±n/a |
|---|
| Temperature | 303.15±n/a K |
|---|
| IC50 | 170±n/a nM |
|---|
| Citation |  Biswal, BK; Morisseau, C; Garen, G; Cherney, MM; Garen, C; Niu, C; Hammock, BD; James, MN The molecular structure of epoxide hydrolase B from Mycobacterium tuberculosis and its complex with a urea-based inhibitor. J Mol Biol381:897-912 (2008) [PubMed] Article Biswal, BK; Morisseau, C; Garen, G; Cherney, MM; Garen, C; Niu, C; Hammock, BD; James, MN The molecular structure of epoxide hydrolase B from Mycobacterium tuberculosis and its complex with a urea-based inhibitor. J Mol Biol381:897-912 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Inhibition_Run data, Solution Info, Assay Method |
|---|
| |
| Epoxide hydrolase B |
|---|
| Name: | Epoxide hydrolase B |
|---|
| Synonyms: | EPHB_MYCTO | Epoxide Hydrolase B (EHB) | Mtb EHB |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 39285.78 |
|---|
| Organism: | Mycobacterium tuberculosis |
|---|
| Description: | The His6-tagged Mtb EHB was expressed in E. coli and purified. |
|---|
| Residue: | 356 |
|---|
| Sequence: | MSQVHRILNCRGTRIHAVADSPPDQQGPLVVLLHGFPESWYSWRHQIPALAGAGYRVVAI
DQRGYGRSSKYRVQKAYRIKELVGDVVGVLDSYGAEQAFVVGHDWGAPVAWTFAWLHPDR
CAGVVGISVPFAGRGVIGLPGSPFGERRPSDYHLELAGPGRVWYQDYFAVQDGIITEIEE
DLRGWLLGLTYTVSGEGMMAATKAAVDAGVDLESMDPIDVIRAGPLCMAEGARLKDAFVY
PETMPAWFTEADLDFYTGEFERSGFGGPLSFYHNIDNDWHDLADQQGKPLTPPALFIGGQ
YDVGTIWGAQAIERAHEVMPNYRGTHMIADVGHWIQQEAPEETNRLLLDFLGGLRP
|
|
|
|---|
| BDBM25728 |
|---|
| BDBM25726 |
|---|
| Name | BDBM25728 |
|---|
| Synonyms: | 1,3-bis(4-chlorophenyl)urea | Urea-based compound, 9 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C13H10Cl2N2O |
|---|
| Mol. Mass. | 281.137 |
|---|
| SMILES | Clc1ccc(NC(=O)Nc2ccc(Cl)cc2)cc1 |
|---|
| Structure | 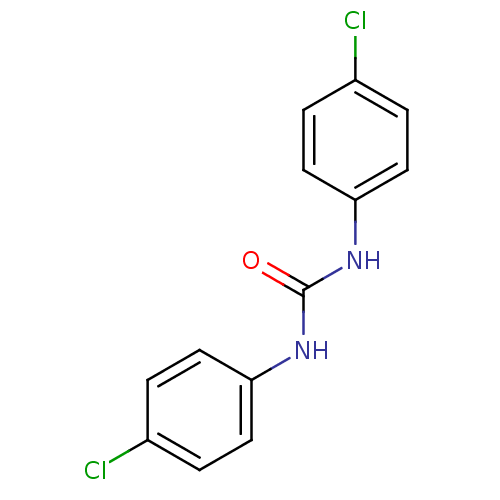
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Biswal, BK; Morisseau, C; Garen, G; Cherney, MM; Garen, C; Niu, C; Hammock, BD; James, MN The molecular structure of epoxide hydrolase B from Mycobacterium tuberculosis and its complex with a urea-based inhibitor. J Mol Biol381:897-912 (2008) [PubMed] Article
Biswal, BK; Morisseau, C; Garen, G; Cherney, MM; Garen, C; Niu, C; Hammock, BD; James, MN The molecular structure of epoxide hydrolase B from Mycobacterium tuberculosis and its complex with a urea-based inhibitor. J Mol Biol381:897-912 (2008) [PubMed] Article