| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Nuclear receptor subfamily 4 group A member 1 |
|---|
| Ligand | BDBM42124 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Fluorescence Polarization Dose Response Assay for TR3-Based Bcl-B Inhibitors |
|---|
| EC50 | 12900±10180 nM |
|---|
| Citation |  PubChem, PC Fluorescence Polarization Dose Response Assay for TR3-Based Bcl-B Inhibitors PubChem Bioassay(2008)[AID] PubChem, PC Fluorescence Polarization Dose Response Assay for TR3-Based Bcl-B Inhibitors PubChem Bioassay(2008)[AID] |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Nuclear receptor subfamily 4 group A member 1 |
|---|
| Name: | Nuclear receptor subfamily 4 group A member 1 |
|---|
| Synonyms: | GFRP1 | HMR | NAK1 | NR4A1 | NR4A1_HUMAN | nuclear receptor subfamily 4, group A, member 1 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 64467.13 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | gi_27894344 |
|---|
| Residue: | 598 |
|---|
| Sequence: | MPCIQAQYGTPAPSPGPRDHLASDPLTPEFIKPTMDLASPEAAPAAPTALPSFSTFMDGY
TGEFDTFLYQLPGTVQPCSSASSSASSTSSSSATSPASASFKFEDFQVYGCYPGPLSGPV
DEALSSSGSDYYGSPCSAPSPSTPSFQPPQLSPWDGSFGHFSPSQTYEGLRAWTEQLPKA
SGPPQPPAFFSFSPPTGPSPSLAQSPLKLFPSQATHQLGEGESYSMPTAFPGLAPTSPHL
EGSGILDTPVTSTKARSGAPGGSEGRCAVCGDNASCQHYGVRTCEGCKGFFKRTVQKNAK
YICLANKDCPVDKRRRNRCQFCRFQKCLAVGMVKEVVRTDSLKGRRGRLPSKPKQPPDAS
PANLLTSLVRAHLDSGPSTAKLDYSKFQELVLPHFGKEDAGDVQQFYDLLSGSLEVIRKW
AEKIPGFAELSPADQDLLLESAFLELFILRLAYRSKPGEGKLIFCSGLVLHRLQCARGFG
DWIDSILAFSRSLHSLLVDVPAFACLSALVLITDRHGLQEPRRVEELQNRIASCLKEHVA
AVAGEPQPASCLSRLLGKLPELRTLCTQGLQRIFYLKLEDLVPPPPIIDKIFMDTLPF
|
|
|
|---|
| BDBM42124 |
|---|
| n/a |
|---|
| Name | BDBM42124 |
|---|
| Synonyms: | 2-(1,3-benzothiazol-2-ylsulfanyl)-N-[(Z)-[1-[(4-methylpiperazin-1-yl)methyl]-2-oxidanylidene-indol-3-ylidene]amino]ethanamide | 2-(1,3-benzothiazol-2-ylsulfanyl)-N-[(Z)-[1-[(4-methylpiperazin-1-yl)methyl]-2-oxoindol-3-ylidene]amino]acetamide | 2-(1,3-benzothiazol-2-ylthio)-N-[(Z)-[1-[(4-methyl-1-piperazinyl)methyl]-2-oxo-3-indolylidene]amino]acetamide | 2-(1,3-benzothiazol-2-ylthio)-N-[(Z)-[2-keto-1-[(4-methylpiperazino)methyl]indolin-3-ylidene]amino]acetamide | BIM-0043480.P001 | cid_5347365 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C23H24N6O2S2 |
|---|
| Mol. Mass. | 480.606 |
|---|
| SMILES | CN1CCN(CN2C(=O)\C(=N/NC(=O)CSc3nc4ccccc4s3)c3ccccc23)CC1 |
|---|
| Structure | 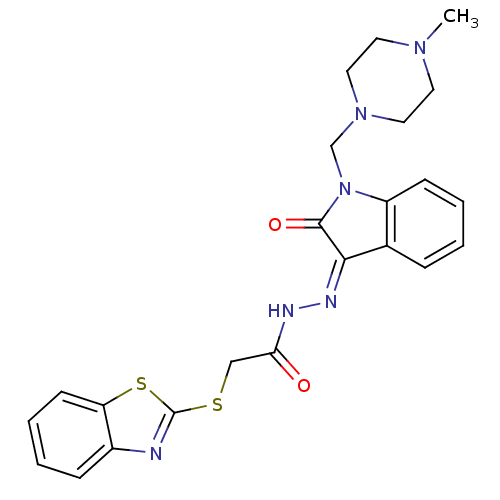
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 PubChem, PC Fluorescence Polarization Dose Response Assay for TR3-Based Bcl-B Inhibitors PubChem Bioassay(2008)[AID]
PubChem, PC Fluorescence Polarization Dose Response Assay for TR3-Based Bcl-B Inhibitors PubChem Bioassay(2008)[AID]