| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Serine/threonine-protein phosphatase PP1-alpha catalytic subunit |
|---|
| Ligand | BDBM64775 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Counterscreen for inhibitors of PP5: fluorescence-based biochemical high throughput dose response assay for inhibitors of Protein Phosphatase 1 (PP1) |
|---|
| IC50 | 67047±n/a nM |
|---|
| Citation |  PubChem, PC Counterscreen for inhibitors of PP5: fluorescence-based biochemical high throughput dose response assay for inhibitors of Protein Phosphatase 1 (PP1) PubChem Bioassay(2010)[AID] PubChem, PC Counterscreen for inhibitors of PP5: fluorescence-based biochemical high throughput dose response assay for inhibitors of Protein Phosphatase 1 (PP1) PubChem Bioassay(2010)[AID] |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Serine/threonine-protein phosphatase PP1-alpha catalytic subunit |
|---|
| Name: | Serine/threonine-protein phosphatase PP1-alpha catalytic subunit |
|---|
| Synonyms: | PP-1A | PP1A_HUMAN | PPP1A | PPP1CA | Serine/threonine protein phosphatase PP1-alpha catalytic subunit | Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | protein phosphatase 1, catalytic subunit, alpha isoform 3 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 37510.66 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_197827 |
|---|
| Residue: | 330 |
|---|
| Sequence: | MSDSEKLNLDSIIGRLLEVQGSRPGKNVQLTENEIRGLCLKSREIFLSQPILLELEAPLK
ICGDIHGQYYDLLRLFEYGGFPPESNYLFLGDYVDRGKQSLETICLLLAYKIKYPENFFL
LRGNHECASINRIYGFYDECKRRYNIKLWKTFTDCFNCLPIAAIVDEKIFCCHGGLSPDL
QSMEQIRRIMRPTDVPDQGLLCDLLWSDPDKDVQGWGENDRGVSFTFGAEVVAKFLHKHD
LDLICRAHQVVEDGYEFFAKRQLVTLFSAPNYCGEFDNAGAMMSVDETLMCSFQILKPAD
KNKGKYGQFSGLNPGGRPITPPRNSAKAKK
|
|
|
|---|
| BDBM64775 |
|---|
| n/a |
|---|
| Name | BDBM64775 |
|---|
| Synonyms: | 5-Amino-4-oxo-3-p-tolyl-3,4-dihydro-thieno[3,4-d]pyridazine-1-carboxylic acid ethyl ester | 5-amino-3-(4-methylphenyl)-4-oxo-1-thieno[3,4-d]pyridazinecarboxylic acid ethyl ester | 5-amino-4-keto-3-(p-tolyl)thieno[3,4-d]pyridazine-1-carboxylic acid ethyl ester | MLS000528230 | SMR000120805 | cid_3153302 | ethyl 5-amino-3-(4-methylphenyl)-4-oxothieno[3,4-d]pyridazine-1-carboxylate | ethyl 5-azanyl-3-(4-methylphenyl)-4-oxidanylidene-thieno[3,4-d]pyridazine-1-carboxylate |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C16H15N3O3S |
|---|
| Mol. Mass. | 329.374 |
|---|
| SMILES | CCOC(=O)c1nn(-c2ccc(C)cc2)c(=O)c2c(N)scc12 |
|---|
| Structure | 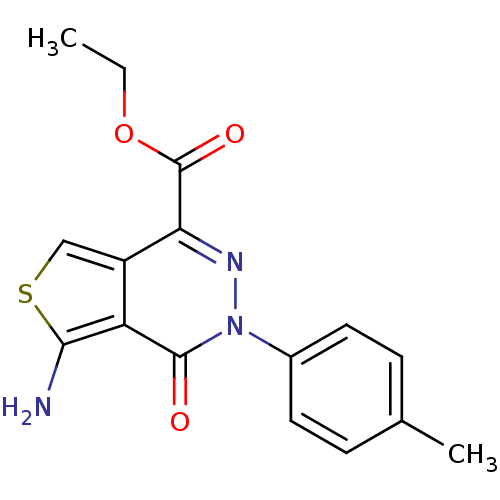
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 PubChem, PC Counterscreen for inhibitors of PP5: fluorescence-based biochemical high throughput dose response assay for inhibitors of Protein Phosphatase 1 (PP1) PubChem Bioassay(2010)[AID]
PubChem, PC Counterscreen for inhibitors of PP5: fluorescence-based biochemical high throughput dose response assay for inhibitors of Protein Phosphatase 1 (PP1) PubChem Bioassay(2010)[AID]