| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Intestinal-type alkaline phosphatase 1 |
|---|
| Ligand | BDBM75626 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Dose Response confirmation of uHTS inhibitors of Human Intestinal Alkaline Phosphatase using Mouse Intestinal Alkaline Phosphatase |
|---|
| IC50 | 29600±n/a nM |
|---|
| Citation |  PubChem, PC Dose Response confirmation of uHTS inhibitors of Human Intestinal Alkaline Phosphatase using Mouse Intestinal Alkaline Phosphatase PubChem Bioassay(2010)[AID] PubChem, PC Dose Response confirmation of uHTS inhibitors of Human Intestinal Alkaline Phosphatase using Mouse Intestinal Alkaline Phosphatase PubChem Bioassay(2010)[AID] |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Intestinal-type alkaline phosphatase 1 |
|---|
| Name: | Intestinal-type alkaline phosphatase 1 |
|---|
| Synonyms: | Alpi | PPBI1_RAT |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 58396.39 |
|---|
| Organism: | Rattus norvegicus (Rat) |
|---|
| Description: | P15693 |
|---|
| Residue: | 540 |
|---|
| Sequence: | MQGDWVLLLLLGLRIHLSFGVIPVEEENPVFWNQKAKEALDVAKKLQPIQTSAKNLILFL
GDGMGVPTVTATRILKGQLGGHLGPETPLAMDHFPFTALSKTYNVDRQVPDSAGTATAYL
CGVKANYKTIGVSAAARFNQCNSTFGNEVFSVMHRAKKAGKSVGVVTTTRVQHASPAGTY
AHTVNRDWYSDADMPSSALQEGCKDIATQLISNMDIDVILGGGRKFMFPKGTPDPEYPGD
SDQSGVRLDSRNLVEEWLAKYQGTRYVWNREQLMQASQDPAVTRLMGLFEPTEMKYDVNR
NASADPSLAEMTEVAVRLLSRNPQGFYLFVEGGRIDQGHHAGTAYLALTEAVMFDSAIEK
ASQLTNEKDTLTLITADHSHVFAFGGYTLRGTSIFGLAPLNAQDGKSYTSILYGNGPGYV
LNSGNRPNVTDAESGDVNYKQQAAVPLSSETHGGEDVAIFARGPQAHLVHGVQEQNYIAH
VMAFAGCLEPYTDCGLAPPADENRPTTPVQNSAITMNNVLLSLQLLVSMLLLVGTALVVS
|
|
|
|---|
| BDBM75626 |
|---|
| n/a |
|---|
| Name | BDBM75626 |
|---|
| Synonyms: | MLS000582497 | N-(3-ethoxypropyl)-4-keto-4-[N'-(2-phenyl-[1,2,4]triazolo[1,5-c]quinazolin-5-yl)hydrazino]butyramide | N-(3-ethoxypropyl)-4-oxidanylidene-4-[2-(2-phenyl-[1,2,4]triazolo[1,5-c]quinazolin-5-yl)hydrazinyl]butanamide | N-(3-ethoxypropyl)-4-oxo-4-[(2-phenyl-[1,2,4]triazolo[1,5-c]quinazolin-5-yl)hydrazo]butanamide | N-(3-ethoxypropyl)-4-oxo-4-[2-(2-phenyl-[1,2,4]triazolo[1,5-c]quinazolin-5-yl)hydrazinyl]butanamide | N-(3-ethoxypropyl)-4-oxo-4-[2-(2-phenyl[1,2,4]triazolo[1,5-c]quinazolin-5-yl)hydrazino]butanamide | SMR000206483 | cid_12004913 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C24H27N7O3 |
|---|
| Mol. Mass. | 461.5163 |
|---|
| SMILES | CCOCCCNC(=O)CCC(=O)NNc1nc2ccccc2c2nc(nn12)-c1ccccc1 |
|---|
| Structure | 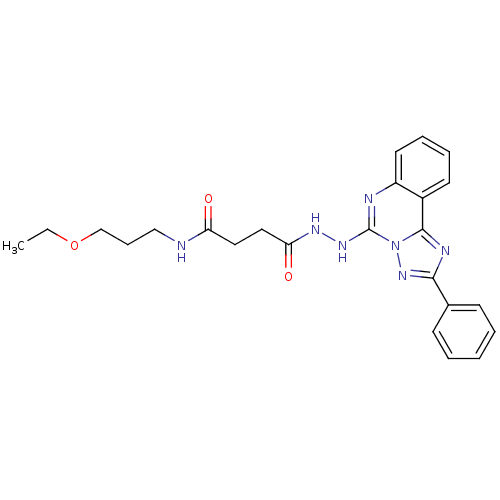
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 PubChem, PC Dose Response confirmation of uHTS inhibitors of Human Intestinal Alkaline Phosphatase using Mouse Intestinal Alkaline Phosphatase PubChem Bioassay(2010)[AID]
PubChem, PC Dose Response confirmation of uHTS inhibitors of Human Intestinal Alkaline Phosphatase using Mouse Intestinal Alkaline Phosphatase PubChem Bioassay(2010)[AID]