| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Cell division protein FtsZ |
|---|
| Ligand | BDBM81332 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Fluorescent Competition Assay |
|---|
| pH | 7.5±0 |
|---|
| IC50 | 1.5e+4±n/a nM |
|---|
| Kd | 15.6±5.7 nM |
|---|
| Citation |  Läppchen, T; Pinas, VA; Hartog, AF; Koomen, GJ; Schaffner-Barbero, C; Andreu, JM; Trambaiolo, D; Löwe, J; Juhem, A; Popov, AV; den Blaauwen, T Probing FtsZ and tubulin with C8-substituted GTP analogs reveals differences in their nucleotide binding sites. Chem Biol15:189-99 (2008) [PubMed] Article Läppchen, T; Pinas, VA; Hartog, AF; Koomen, GJ; Schaffner-Barbero, C; Andreu, JM; Trambaiolo, D; Löwe, J; Juhem, A; Popov, AV; den Blaauwen, T Probing FtsZ and tubulin with C8-substituted GTP analogs reveals differences in their nucleotide binding sites. Chem Biol15:189-99 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Cell division protein FtsZ |
|---|
| Name: | Cell division protein FtsZ |
|---|
| Synonyms: | FTSZ_AQUAE | ftsZ |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 40146.54 |
|---|
| Organism: | Aquifex aeolicus |
|---|
| Description: | O66809 |
|---|
| Residue: | 367 |
|---|
| Sequence: | MEEFVNPCKIKVIGVGGGGSNAVNRMYEDGIEGVELYAINTDVQHLSTLKVPNKIQIGEK
VTRGLGAGAKPEVGEEAALEDIDKIKEILRDTDMVFISAGLGGGTGTGAAPVIAKTAKEM
GILTVAVATLPFRFEGPRKMEKALKGLEKLKESSDAYIVIHNDKIKELSNRTLTIKDAFK
EVDSVLSKAVRGITSIVVTPAVINVDFADVRTTLEEGGLSIIGMGEGRGDEKADIAVEKA
VTSPLLEGNTIEGARRLLVTIWTSEDIPYDIVDEVMERIHSKVHPEAEIIFGAVLEPQEQ
DFIRVAIVATDFPEEKFQVGEKEVKFKVIKKEEKEEPKEEPKPLSDTTYLEEEEIPAVIR
RKNKRLL
|
|
|
|---|
| BDBM81332 |
|---|
| n/a |
|---|
| Name | BDBM81332 |
|---|
| Synonyms: | PyrrGTP (7b) |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C14H20N6O14P3 |
|---|
| Mol. Mass. | 589.2633 |
|---|
| SMILES | Nc1nc2n([C@@H]3O[C@H](COP([O-])(=O)OP([O-])(=O)OP(O)([O-])=O)C(O)C3O)c(nc2c(=O)[nH]1)N1CCCC1 |r| |
|---|
| Structure | 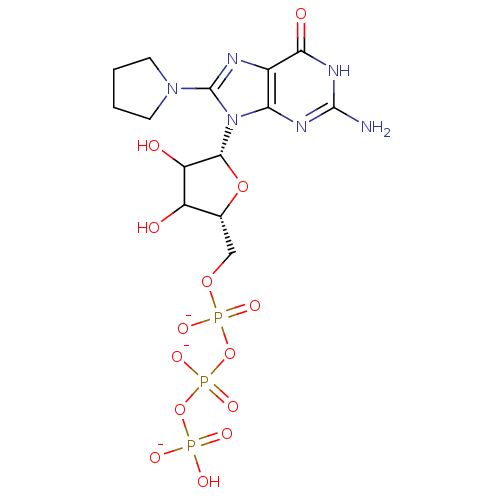
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Läppchen, T; Pinas, VA; Hartog, AF; Koomen, GJ; Schaffner-Barbero, C; Andreu, JM; Trambaiolo, D; Löwe, J; Juhem, A; Popov, AV; den Blaauwen, T Probing FtsZ and tubulin with C8-substituted GTP analogs reveals differences in their nucleotide binding sites. Chem Biol15:189-99 (2008) [PubMed] Article
Läppchen, T; Pinas, VA; Hartog, AF; Koomen, GJ; Schaffner-Barbero, C; Andreu, JM; Trambaiolo, D; Löwe, J; Juhem, A; Popov, AV; den Blaauwen, T Probing FtsZ and tubulin with C8-substituted GTP analogs reveals differences in their nucleotide binding sites. Chem Biol15:189-99 (2008) [PubMed] Article