| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Alpha-1D adrenergic receptor |
|---|
| Ligand | BDBM50263488 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEBML_1698006 |
|---|
| Ki | 91±n/a nM |
|---|
| Citation |  Del Bello, F; Bonifazi, A; Giorgioni, G; Cifani, C; Micioni Di Bonaventura, MV; Petrelli, R; Piergentili, A; Fontana, S; Mammoli, V; Yano, H; Matucci, R; Vistoli, G; Quaglia, W 1-[3-(4-Butylpiperidin-1-yl)propyl]-1,2,3,4-tetrahydroquinolin-2-one (77-LH-28-1) as a Model for the Rational Design of a Novel Class of Brain Penetrant Ligands with High Affinity and Selectivity for Dopamine D J Med Chem61:3712-3725 (2018) [PubMed] Article Del Bello, F; Bonifazi, A; Giorgioni, G; Cifani, C; Micioni Di Bonaventura, MV; Petrelli, R; Piergentili, A; Fontana, S; Mammoli, V; Yano, H; Matucci, R; Vistoli, G; Quaglia, W 1-[3-(4-Butylpiperidin-1-yl)propyl]-1,2,3,4-tetrahydroquinolin-2-one (77-LH-28-1) as a Model for the Rational Design of a Novel Class of Brain Penetrant Ligands with High Affinity and Selectivity for Dopamine D J Med Chem61:3712-3725 (2018) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Alpha-1D adrenergic receptor |
|---|
| Name: | Alpha-1D adrenergic receptor |
|---|
| Synonyms: | ADA1D_HUMAN | ADRA1A | ADRA1D | Adrenergic receptor | Adrenergic receptor alpha | Alpha 1D-adrenoceptor | Alpha 1D-adrenoreceptor | Alpha adrenergic receptor (1a and 1d) | Alpha-1D adrenoceptor | Alpha-adrenergic receptor 1a | adrenergic Alpha1D |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 60485.82 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | adrenergic Alpha1D ADRA1D HUMAN::P25100 |
|---|
| Residue: | 572 |
|---|
| Sequence: | MTFRDLLSVSFEGPRPDSSAGGSSAGGGGGSAGGAAPSEGPAVGGVPGGAGGGGGVVGAG
SGEDNRSSAGEPGSAGAGGDVNGTAAVGGLVVSAQGVGVGVFLAAFILMAVAGNLLVILS
VACNRHLQTVTNYFIVNLAVADLLLSATVLPFSATMEVLGFWAFGRAFCDVWAAVDVLCC
TASILSLCTISVDRYVGVRHSLKYPAIMTERKAAAILALLWVVALVVSVGPLLGWKEPVP
PDERFCGITEEAGYAVFSSVCSFYLPMAVIVVMYCRVYVVARSTTRSLEAGVKRERGKAS
EVVLRIHCRGAATGADGAHGMRSAKGHTFRSSLSVRLLKFSREKKAAKTLAIVVGVFVLC
WFPFFFVLPLGSLFPQLKPSEGVFKVIFWLGYFNSCVNPLIYPCSSREFKRAFLRLLRCQ
CRRRRRRRPLWRVYGHHWRASTSGLRQDCAPSSGDAPPGAPLALTALPDPDPEPPGTPEM
QAPVASRRKPPSAFREWRLLGPFRRPTTQLRAKVSSLSHKIRAGGAQRAEAACAQRSEVE
AVSLGVPHEVAEGATCQAYELADYSNLRETDI
|
|
|
|---|
| BDBM50263488 |
|---|
| n/a |
|---|
| Name | BDBM50263488 |
|---|
| Synonyms: | CHEMBL4085780 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H27N3O |
|---|
| Mol. Mass. | 349.4693 |
|---|
| SMILES | O=C1CCc2ccccc2N1CCCN1CCN(CC1)c1ccccc1 |
|---|
| Structure | 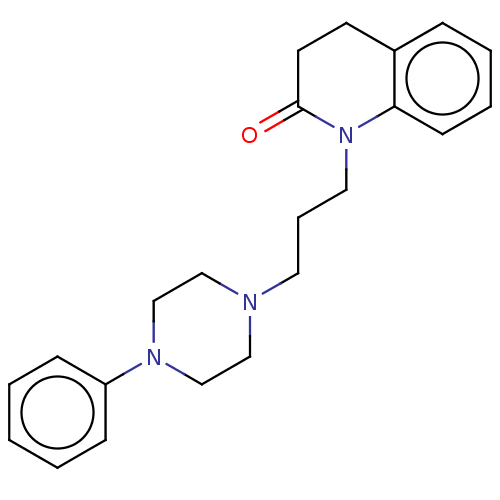
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Del Bello, F; Bonifazi, A; Giorgioni, G; Cifani, C; Micioni Di Bonaventura, MV; Petrelli, R; Piergentili, A; Fontana, S; Mammoli, V; Yano, H; Matucci, R; Vistoli, G; Quaglia, W 1-[3-(4-Butylpiperidin-1-yl)propyl]-1,2,3,4-tetrahydroquinolin-2-one (77-LH-28-1) as a Model for the Rational Design of a Novel Class of Brain Penetrant Ligands with High Affinity and Selectivity for Dopamine D J Med Chem61:3712-3725 (2018) [PubMed] Article
Del Bello, F; Bonifazi, A; Giorgioni, G; Cifani, C; Micioni Di Bonaventura, MV; Petrelli, R; Piergentili, A; Fontana, S; Mammoli, V; Yano, H; Matucci, R; Vistoli, G; Quaglia, W 1-[3-(4-Butylpiperidin-1-yl)propyl]-1,2,3,4-tetrahydroquinolin-2-one (77-LH-28-1) as a Model for the Rational Design of a Novel Class of Brain Penetrant Ligands with High Affinity and Selectivity for Dopamine D J Med Chem61:3712-3725 (2018) [PubMed] Article