

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Bifunctional epoxide hydrolase 2 | ||
| Ligand | BDBM50454626 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1752865 (CHEMBL4187625) | ||
| IC50 | 4.0±n/a nM | ||
| Citation |  Kodani, SD; Bhakta, S; Hwang, SH; Pakhomova, S; Newcomer, ME; Morisseau, C; Hammock, BD Identification and optimization of soluble epoxide hydrolase inhibitors with dual potency towards fatty acid amide hydrolase. Bioorg Med Chem Lett28:762-768 (2018) [PubMed] Article Kodani, SD; Bhakta, S; Hwang, SH; Pakhomova, S; Newcomer, ME; Morisseau, C; Hammock, BD Identification and optimization of soluble epoxide hydrolase inhibitors with dual potency towards fatty acid amide hydrolase. Bioorg Med Chem Lett28:762-768 (2018) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Bifunctional epoxide hydrolase 2 | |||
| Name: | Bifunctional epoxide hydrolase 2 | ||
| Synonyms: | Cytosolic epoxide hydrolase 2 | EBifunctional epoxide hydrolase 2 | EPHX2 | Epoxide hydratase | HYES_HUMAN | Lipid-phosphate phosphatase | Soluble epoxide hydrolase (sEH) | epoxide hydrolase 2, cytoplasmic | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 62613.07 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P34913 | ||
| Residue: | 555 | ||
| Sequence: |
| ||
| BDBM50454626 | |||
| n/a | |||
| Name | BDBM50454626 | ||
| Synonyms: | CHEMBL4204023 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C21H21F3N2O4 | ||
| Mol. Mass. | 422.3976 | ||
| SMILES | FC(F)(F)Oc1ccc(NC(=O)N[C@H]2CC[C@@H](CC2)Oc2ccc(C=O)cc2)cc1 |r,wU:16.19,wD:13.12,(30.72,-33.27,;30.72,-34.67,;31.96,-33.65,;29.47,-33.65,;30.72,-36.21,;32.05,-36.98,;32.05,-38.52,;33.38,-39.29,;34.72,-38.52,;36.06,-39.29,;37.39,-38.52,;37.39,-36.98,;38.72,-39.28,;40.06,-38.51,;41.38,-39.29,;42.71,-38.52,;42.72,-36.98,;41.38,-36.21,;40.04,-36.98,;44.05,-36.22,;45.39,-36.99,;45.38,-38.53,;46.71,-39.3,;48.04,-38.53,;49.38,-39.3,;49.38,-40.84,;48.04,-36.98,;46.71,-36.22,;34.72,-36.97,;33.38,-36.21,)| | ||
| Structure | 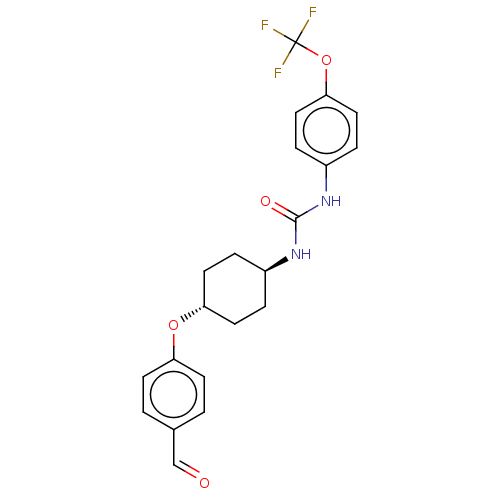
| ||